|
|
| With microbes it doesn't matter who they are, but what they do |
| By Stilianos Louca. December 5, 2016 |
Introduction
One of the challenges of understanding natural microbial systems is that they are incredibly complex.
A gram of soil, for example, contains millions of microorganisms and thousands of microbial species.
Despite this enormous diversity, most major biochemical reactions in natural ecosystems are driven by a relatively small number of metabolic "pathways" (i.e., chains of one or more chemical reactions), which microorganisms use to exploit the various energy sources available.
Such pathways include photosynthesis (the use of light energy), methanotrophy (consuming methane) or heterotrophic sulfate respiration (breathing sulfate when oxygen is scarce).
These pathways can be performed by a multitude of distinct microbial species, often coexisting in the same environment.
The exact relationship between the species composition of a microbial system and the metabolic functions performed in that system is poorly understood, and this impedes our ability to predict the biochemical processes occurring in nature.
We do know however that the growth of microorganisms is influenced by environmental conditions, which constrain which metabolic pathways could in principle provide sufficient energy for growth (for example photosynthesis is impossible in the dark).
One could thus hypothesize that environmental conditions may fully prescribe the distribution (and perhaps even abundance) of organisms utilizing specific metabolic pathways, regardless of which particular species happens to perform each pathway.
If this hypothesis turns out to be correct, it would greatly simplify our understanding of microbially driven biochemical processes in natural ecosystems.
Examining the species-function relationship in a natural miniature ecosystem
In our latest study published in the journal Nature Ecology & Evolution, we tested the above hypothesis by examining the relationship between the species composition of a natural microbial system and its capacity to perform various metabolic functions.
For this task, we needed some type of natural ecosystem that's easily replicated, easy to survey and able to sustain multiple microbial species. Bromeliad plants of the species Aechmea nudicaulis have natural cavities formed by their foliage, and these cavities seemed like a good choice for us:
In some regions of the world (such as the Jurubatiba National Park in Brazil), these bromeliads occur in high numbers, and their tanks fill with rainwater and detritus, the decomposition of which supports rich microbial communities.
As some of our colleagues from Canada and Brazil had extensive experience using bromeliads as experimental systems for ecology, this seemed like (and turned out to be) a perfect collaboration!
Field work in Brazil went through rather smoothly and without any grand surprises, apart from a couple of snakes and frogs occasionally encountered inside the bromeliads.
We used DNA sequencing to identify which microbial species lived in each bromeliad as well as what types of metabolic functions are present and to what extent.
Samples for DNA analysis were immediately flash-frozen in liquid nitrogen (which is colder than -195 °C), in a "high-tech" setup involving socks attached to strings - where there's a will there's a solution in the Brazilian bush.
To put things into an environmental context, we also took a multitude of physical and chemical measurements in and around each bromeliad.
I must admit that I've learned a lot from this excursion; for example that even the toughest hiking boots dissolve in hot Brazilian sand-dunes, and that cheap centrifuge tubes explode violently when taken out of liquid nitrogen.
Alternative species end up performing the same metabolic pathways
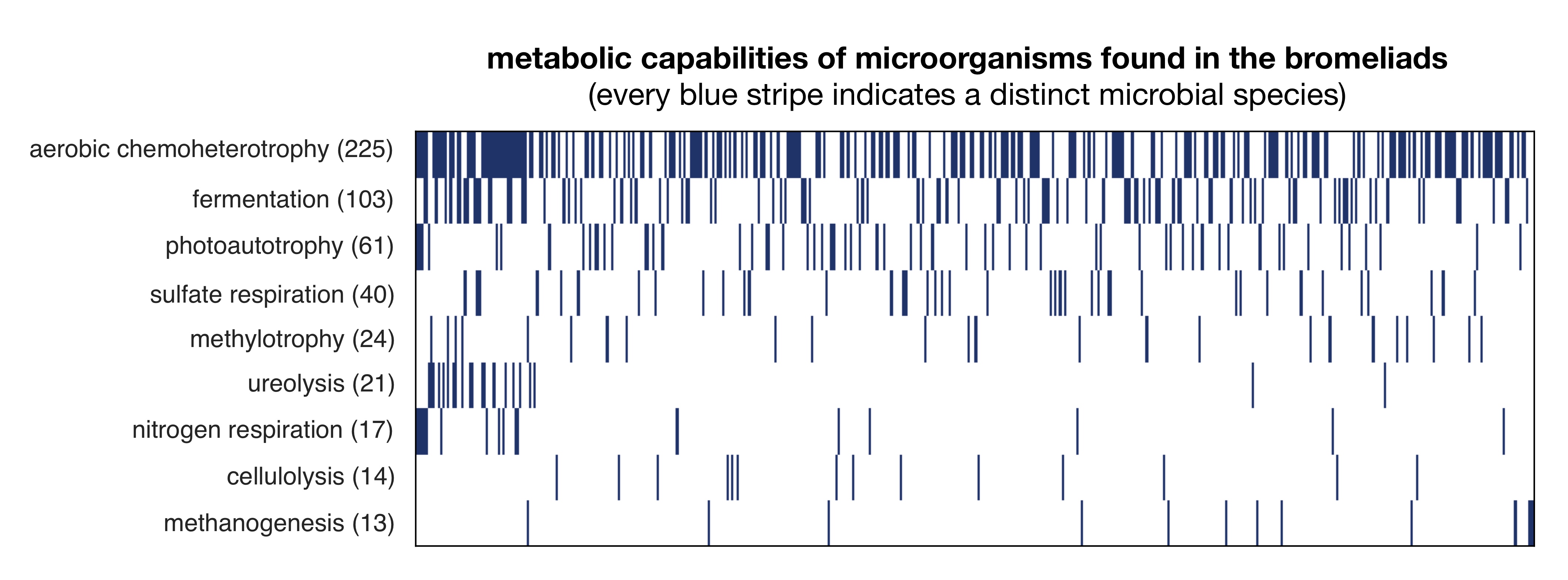
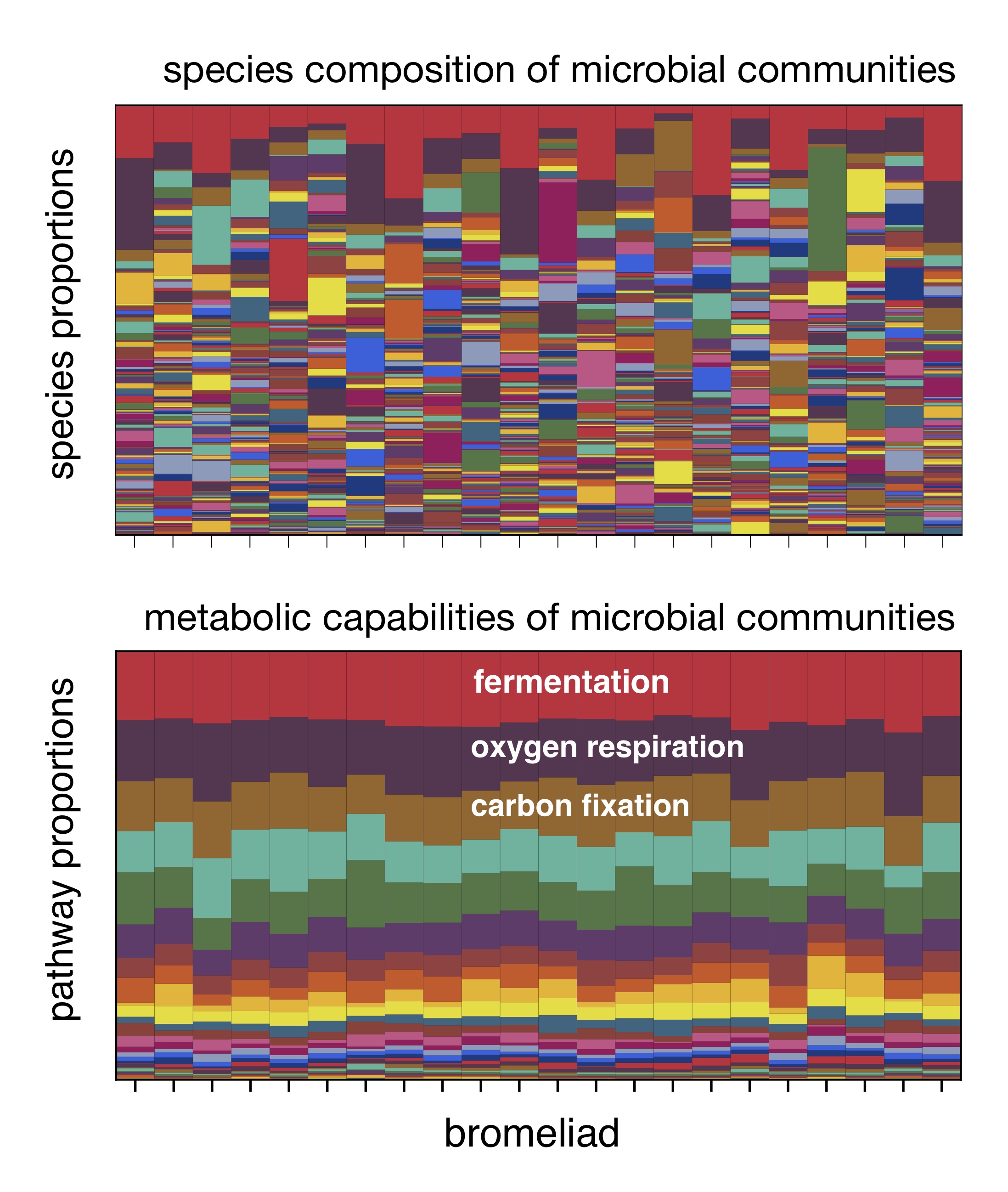
When we analyzed the DNA of the surveyed microorganisms, we found that microorganisms exhibited remarkably similar metabolic capabilities in all bromeliads, when measured in terms of the proportions of genes responsible for various metabolic pathways.
In contrast, the species composition was drastically different in each bromeliad, which meant that the same metabolic pathways were performed by completely different species each time.
For example, when combined across bromeliads, we found dozens of different photosynthetic bacteria, but in each bromeliad only a small subset of these bacteria were present.
This finding was even more remarkable in view of the fact that all of the sampled bromeliads were only a few dozen meters apart from one another, and hence one would have expected that roughly the same microbial species would colonize each bromeliad.
Our findings showed that even adjacent ecosystems with similar conditions can harbor very distinct microbial species.
Furthermore, even if microbial communities differ between systems, this need not imply differences in the actual biochemical processes performed.
If environmental conditions strongly influence the metabolic activity of microbial systems, but exert less influence on who gets to perform each metabolic function, then what could possibly cause this intense variation of species across systems?
Using plenty of stats, we concluded that this variation may have been caused by intrinsic biological interactions between microorganisms, such as predation by viruses infecting bacteria.
Conclusions
It still remains unresolved what exactly drives and controls this variation of microbial species despite a strongly conserved biochemistry.
Whatever the answer may be, our work has shown that it is important to differentiate between variations of microbial species and variations of biochemical functions, for example when assessing the stability/health of an ecosystem or when evaluating the effects of medical treatments on the human gut microbiota.
When it comes to microbes, it seems to be more important to know what they do, rather than who they are.
Full scientific article:
Louca, S., Jacques, S. M. S., Pires, A. P. F., Leal, J. S., Srivastava, D. S., Parfrey, L. W., Farjalla, V. F., Doebeli, M. (2016). High taxonomic variability despite stable functional structure across microbial communities. Nature Ecology & Evolution 1:0015
|
|
 ×
 ×
 ×
 ×
|
|
|
Louca lab. Department of Biology, University of Oregon, Eugene, USA
© 2024 Stilianos Louca all rights reserved
|
|
|



