| Bromeliad microbiomes, Brazil |
|
|
 |
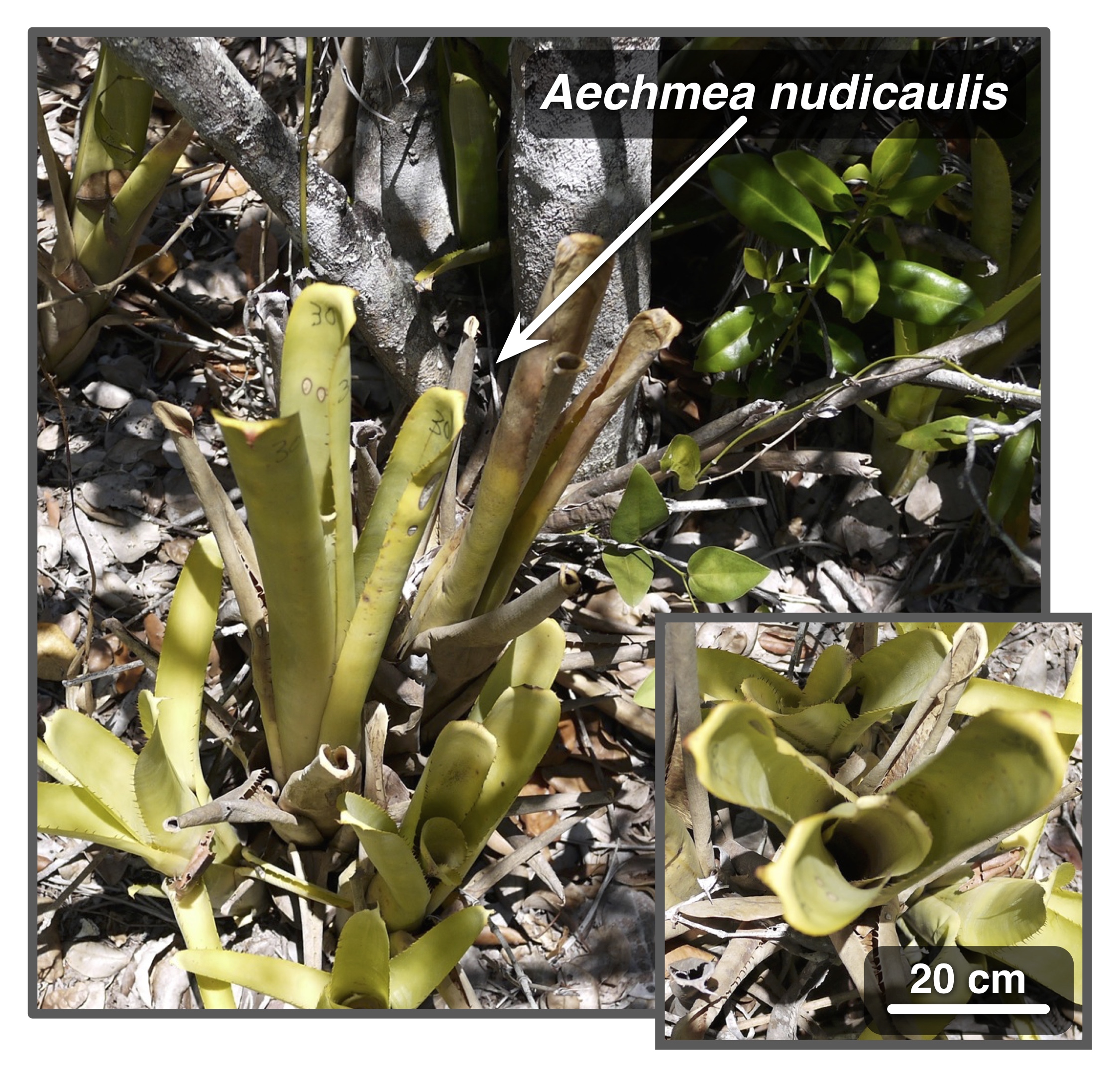
| Data from our study on the taxonomic and functional structure of microbial communities in bromeliad plants, in Jurubatiba National Park, Brazil. 16S rRNA amplicon sequencing |
|
| Reference: Louca, S., Jacques, S. M. S., Pires, A. P. F., Leal, J. S., Srivastava, D. S., Parfrey, L. W., Farjalla, V. F., Doebeli, M. (2016). High taxonomic variability despite stable functional structure across microbial communities. Nature Ecology & Evolution 1:0015 |
|
×
|
|
|
| Global Prokaryotic Census (GPC) |
|
|
 |
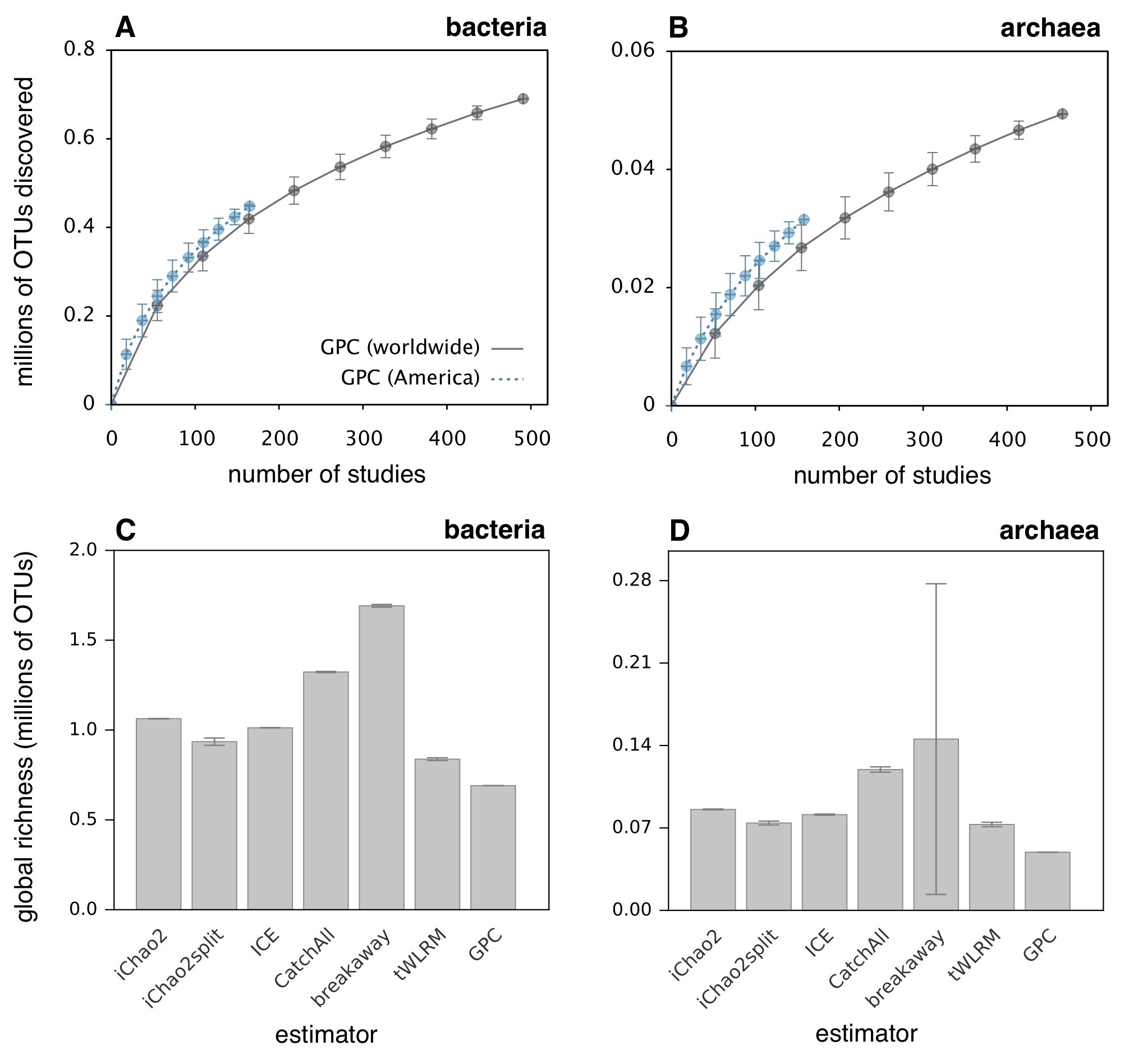
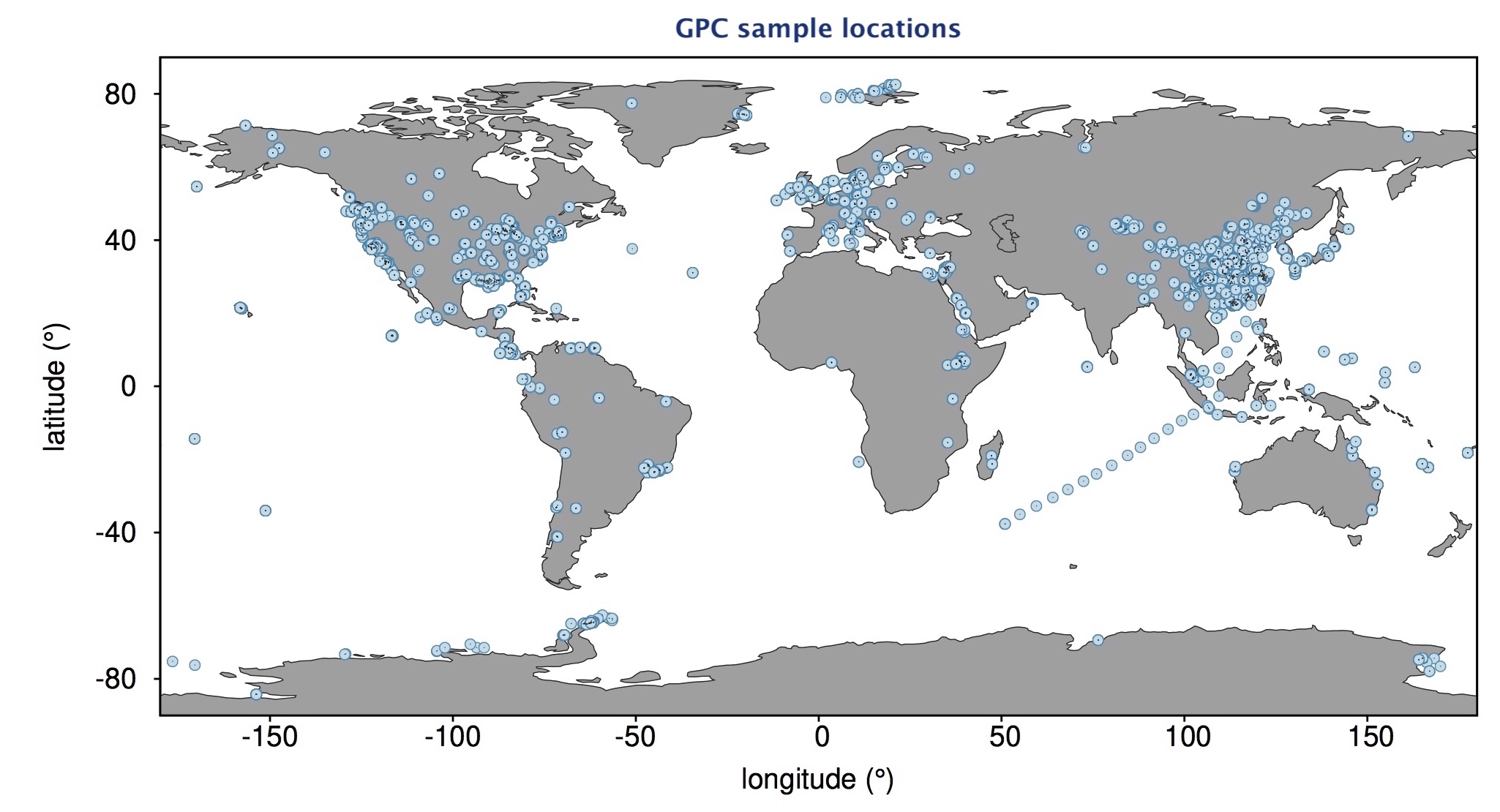
| A massive composite dataset of bacterial and archaeal 16S amplicon sequences, compiled from ~34,000 samples across 492 studies, covering a multitude of environments worldwide. |
|
| Reference: Louca, S., Mazel, F., Doebeli, M., Parfrey, W. L. (2019). A census-based estimate of Earth's bacterial and archaeal diversity. PLOS Biology 17:e3000106 |
|
×
×
|
|
|
| Challenge data for macroevolutionary reconstructions |
|
|
 |
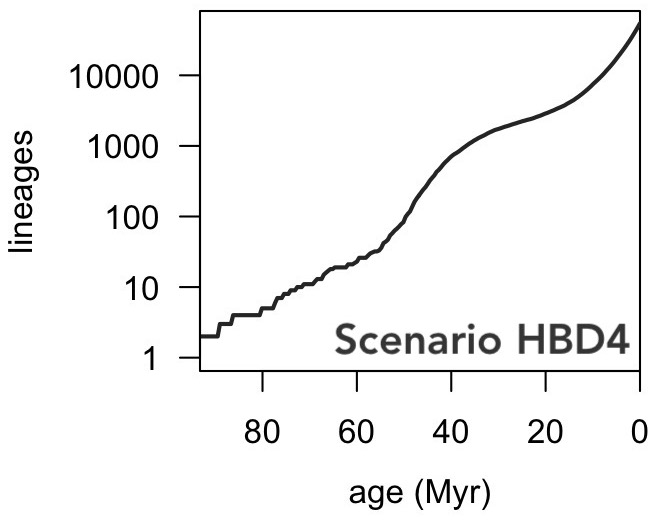
| Challenge data that demonstrates the difficulties of reconstructing speciation/extinction dynamics over time from extant timetrees. |
|
| Reference: Louca, S., Pennell, M.W. (2020). Extant timetrees are consistent with a myriad of diversification histories. Nature 580:502-505 |
|
×
|
|
|
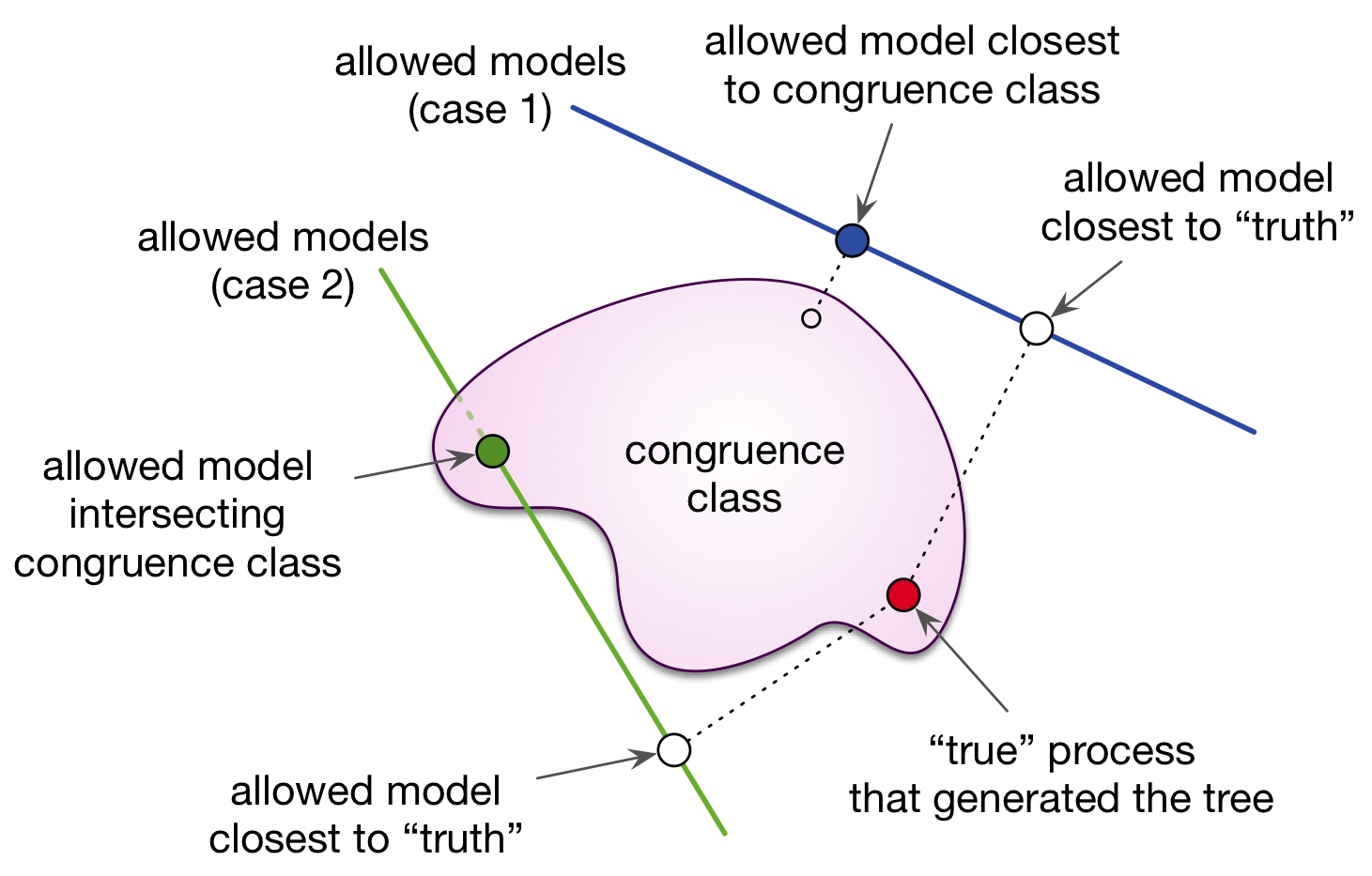
| Model congruency in macroevolution |
|
|
 |
| Supporting data and code for our paper on congruence classes in macroevolutionary birth-death models and on pulled diversification rates. |
|
| Reference: Louca, S., Pennell, M.W. (2020). Extant timetrees are consistent with a myriad of diversification histories. Nature 580:502-505 |
|
×
|
|
|
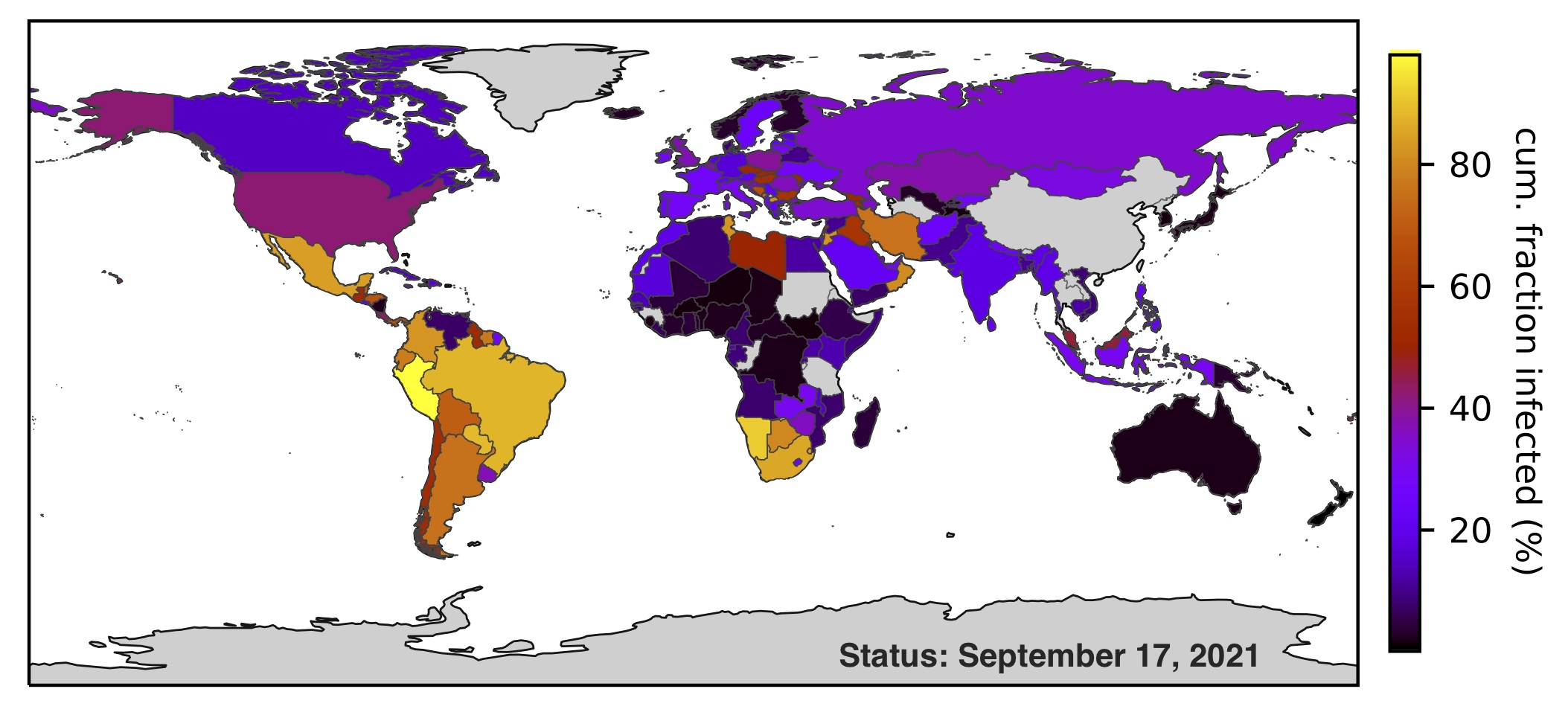
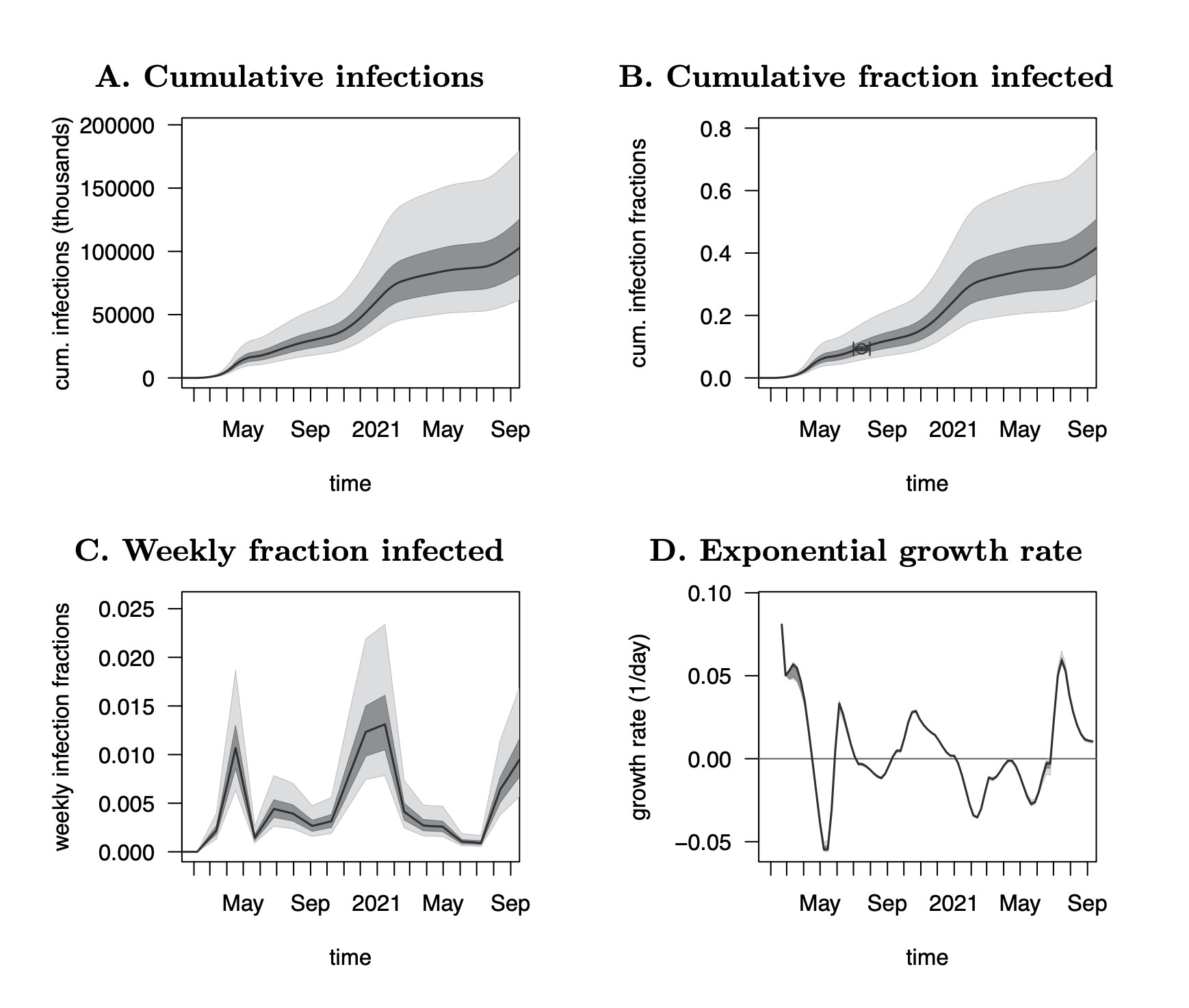
| SARS-CoV-2 infections across countries and over time |
|
|
 |
| Estimation of weekly and cumulative SARS-CoV-2 infection counts in 165 countries over time. |
|
| Reference: Louca, S. (2021). SARS-CoV-2 infections in 165 countries over time. International Journal of Infectious Diseases 111:336-346 |
|
×
×
|
|
|
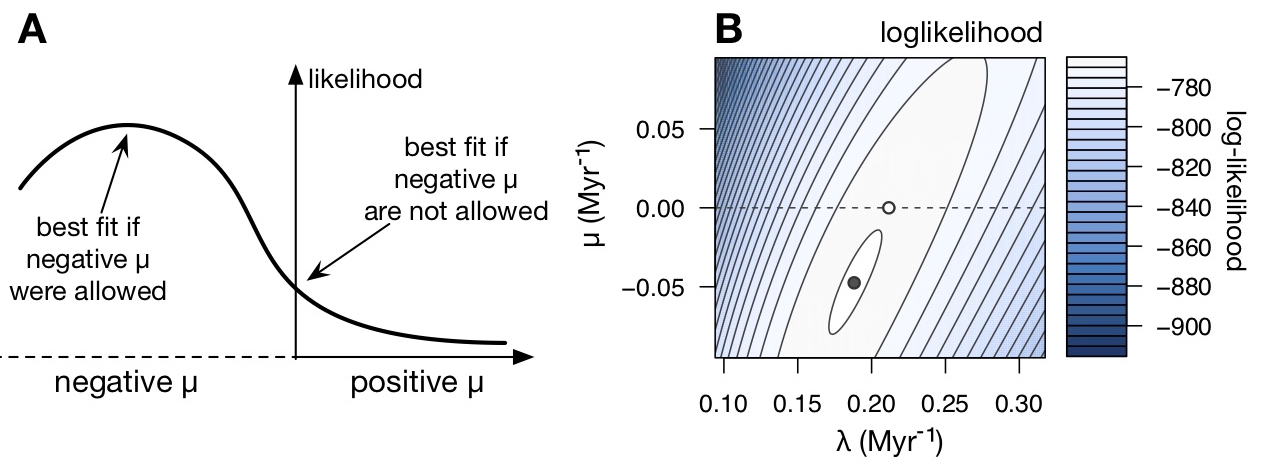
| Zero extinction rate estimates |
|
|
 |
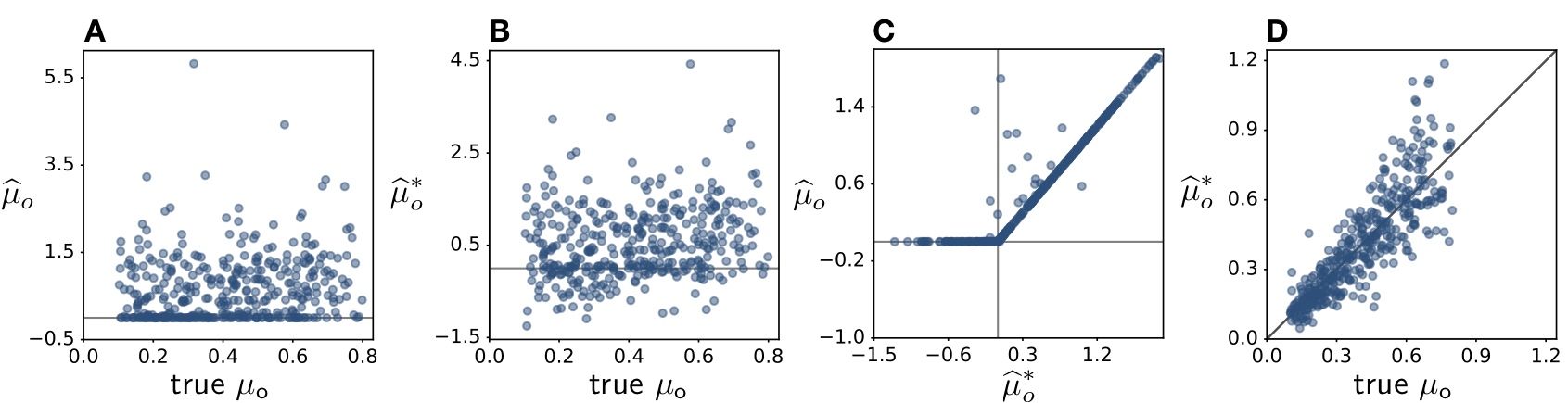
| Supporting data and code for our manuscript on zero extinction rate estimates from phylogenies. |
|
| Reference: Louca, S., Pennell, M.W. (2021). Why extinction estimates from extant phylogenies are so often zero. Current Biology 31:3168-3173 |
|
×
×
|
|
|
| Delineating marine microbial taxa |
|
|
 |
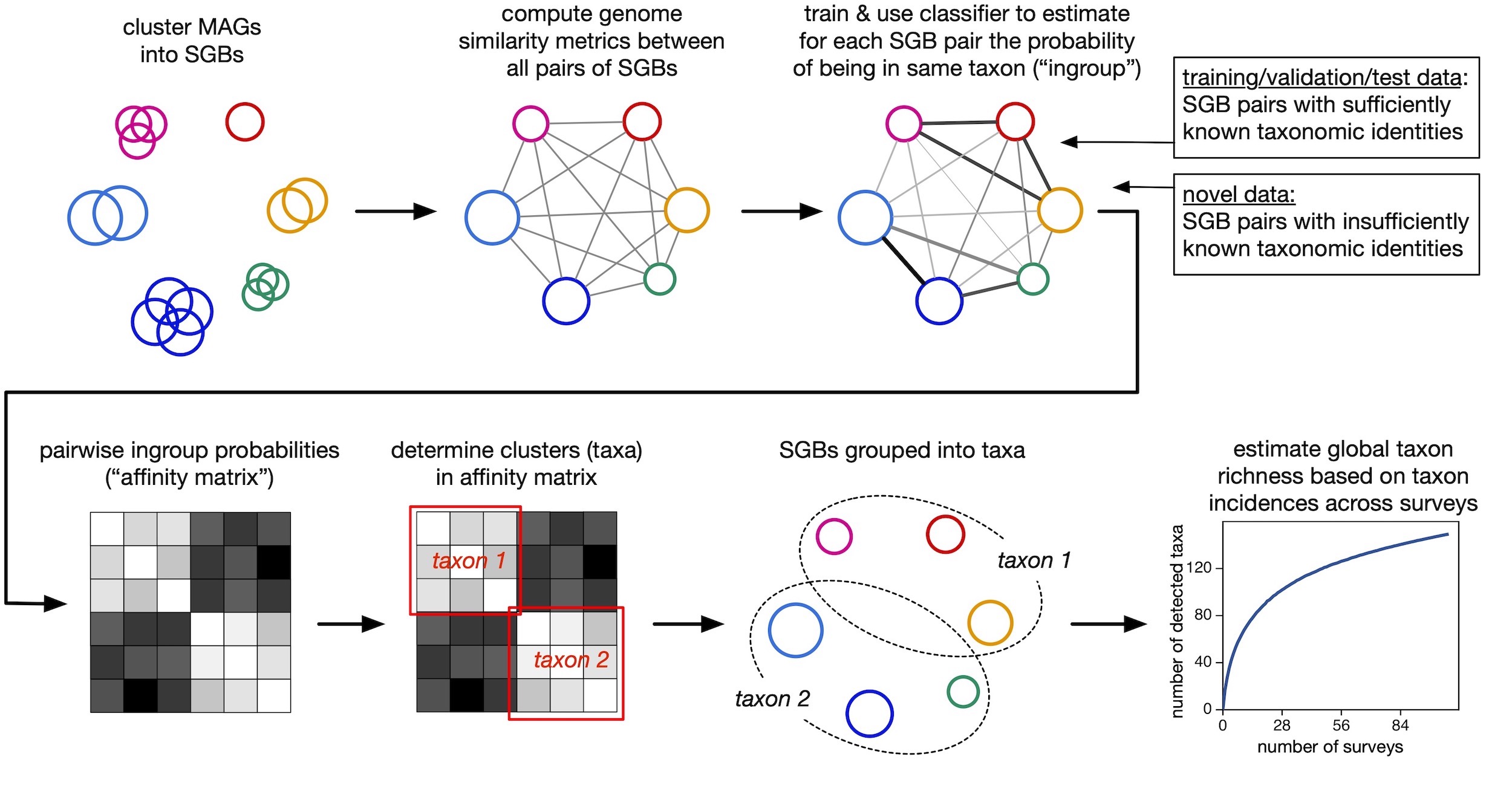
| Supporting data and code for our paper on delineating marine microbial taxa using machine learning models and genome similarity metrics. |
|
×
|
|
|
| Big and Little Soda Lake microbiomes, Nevada |
|
|
 |
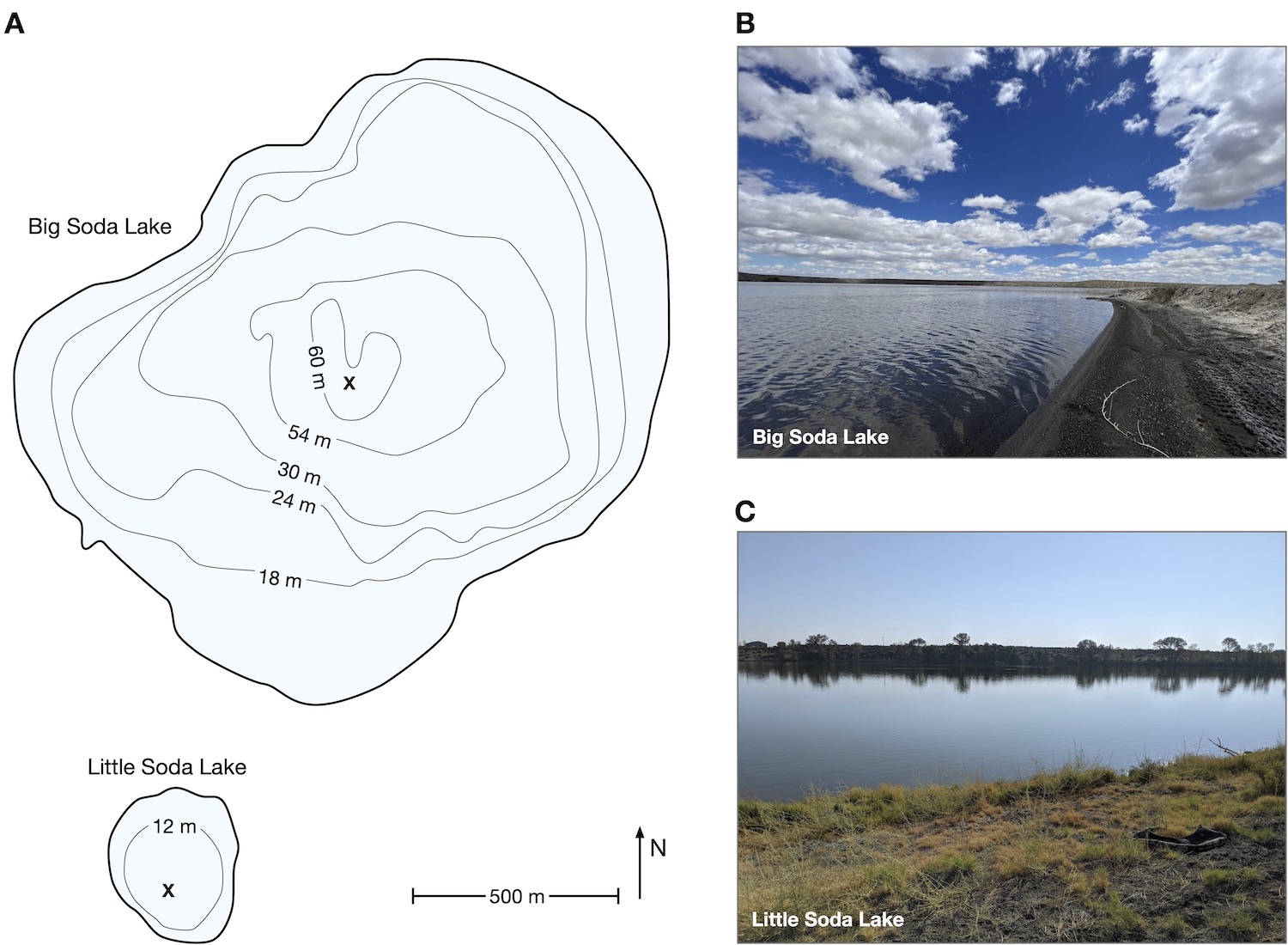
| Data from our microbiome survey of Big and Little Soda Lakes in Navada, USA. |
|
| Reference: Habibi-Soufi, H., Tran, D., Louca, S. (2024). Microbiology of Big Soda Lake, a multi-extreme meromictic volcanic crater lake in the Nevada desert. Environmental Microbiology 26:e16578 |
|
×
|
|
|



