|
|
| |
| Software developed by the Louca lab
|
|
| |
| Audio processing |
|
|
 |
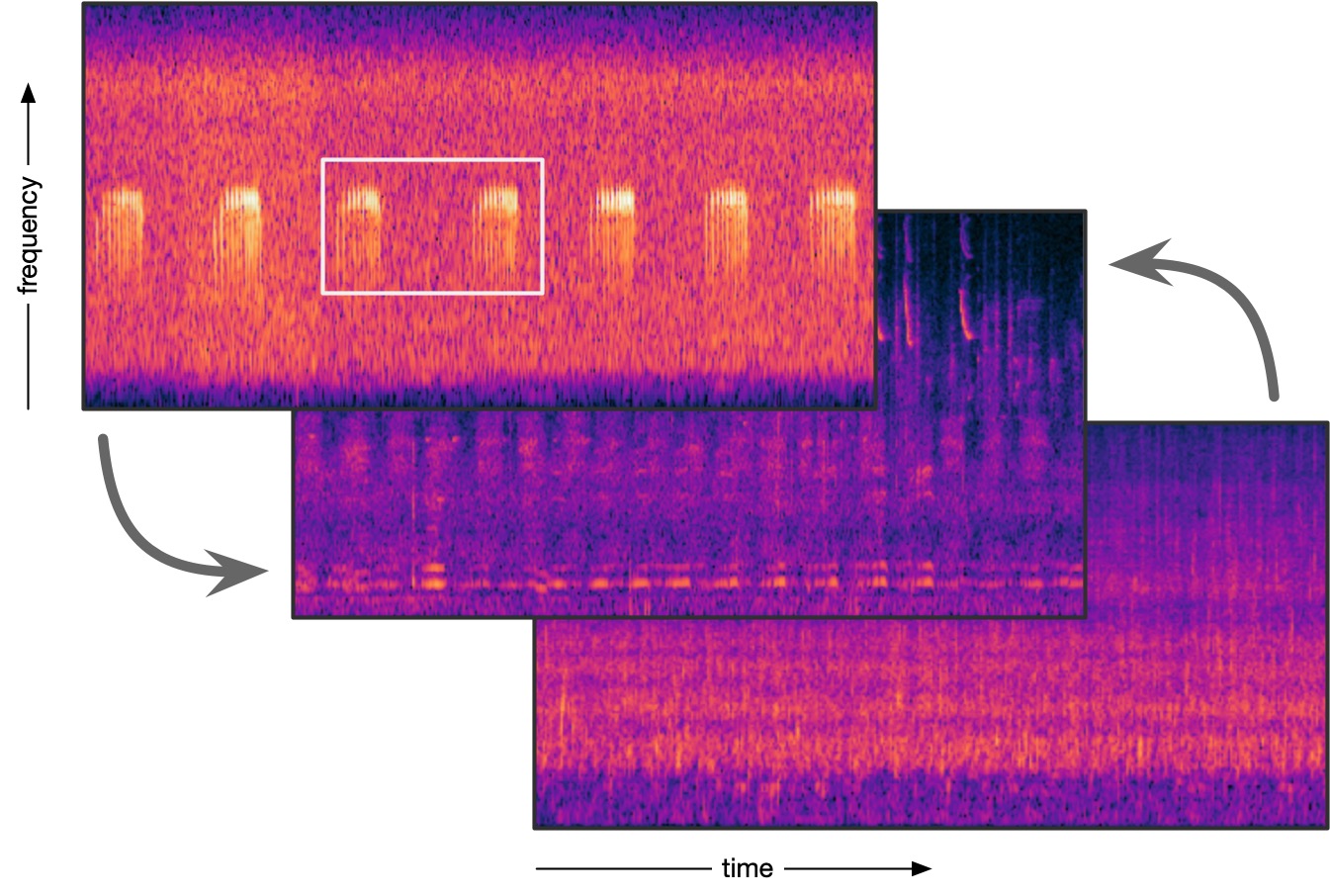
| Python scripts for working with bioacoustic audio files (WAV or FLAC), such as extracting clips, converting annotation tables, plotting spectrograms and computing acoustic parameter differences. |
|
×
|
|
|
| Cariaco metabolic modeling |
|
|
 |
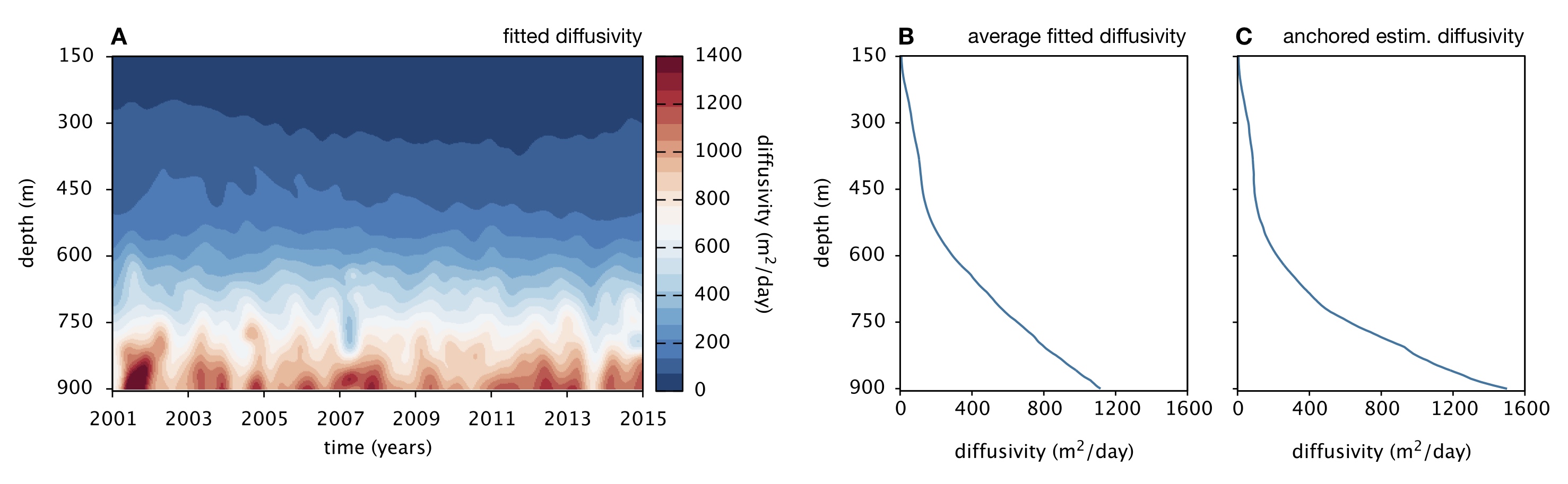
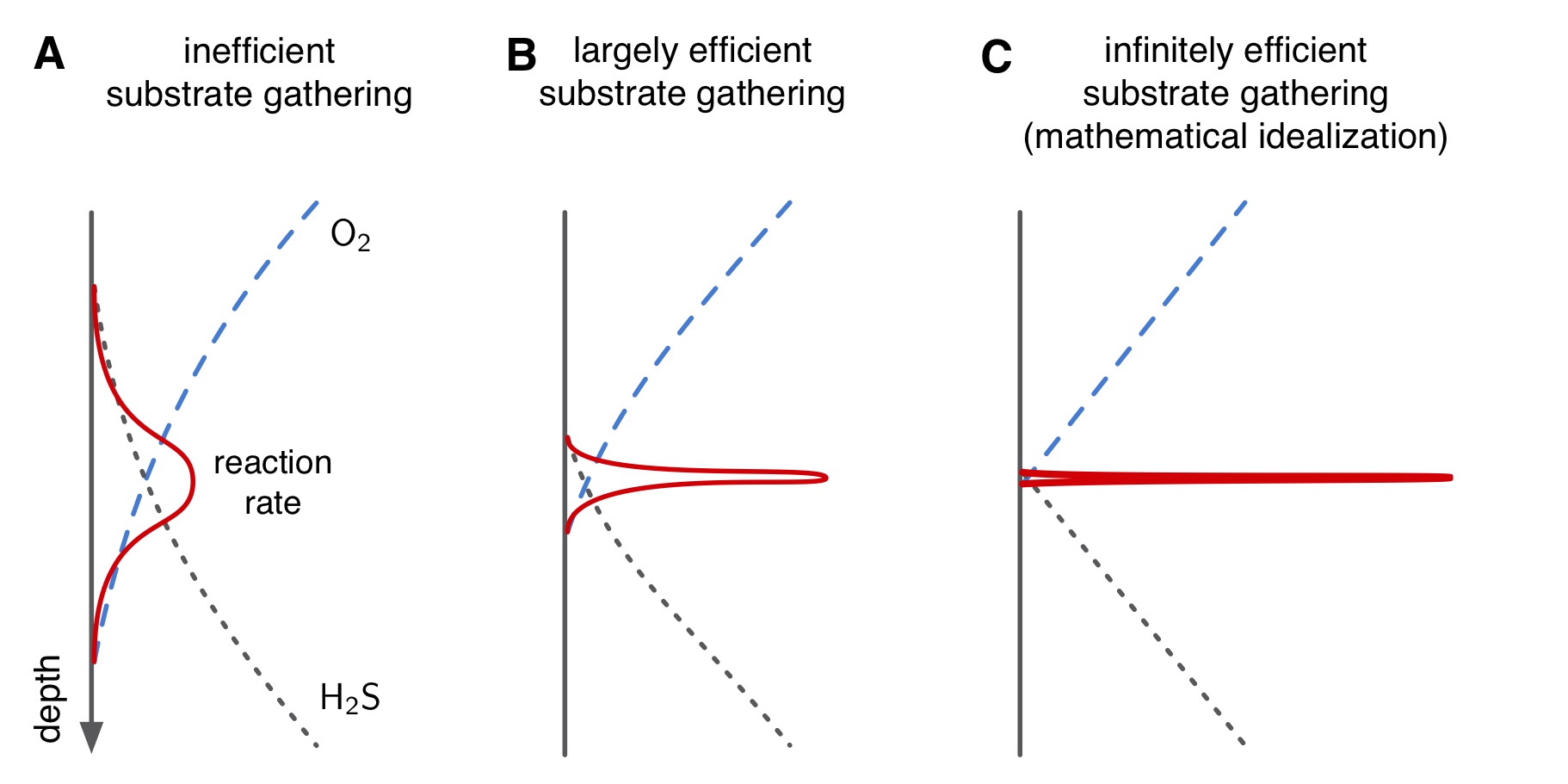
| MATLAB code for inverse linear transport modeling (ILTM), estimation of eddy diffusivity based on the spatiotemporal distribution of a conserved tracer, and spatial metabolic flux (SMF) analysis of microbial systems near steady state, exemplified for the Cariaco Basin sub-euphotic water column. |
|
| Reference: Louca, S., Scranton, M. I., Taylor, G. T., Astor, Y. M., Crowe, S. A., Doebeli, M. (2019). Circumventing kinetics in biogeochemical modeling. PNAS 116:11329-11338 |
|
×
×
|
|
|
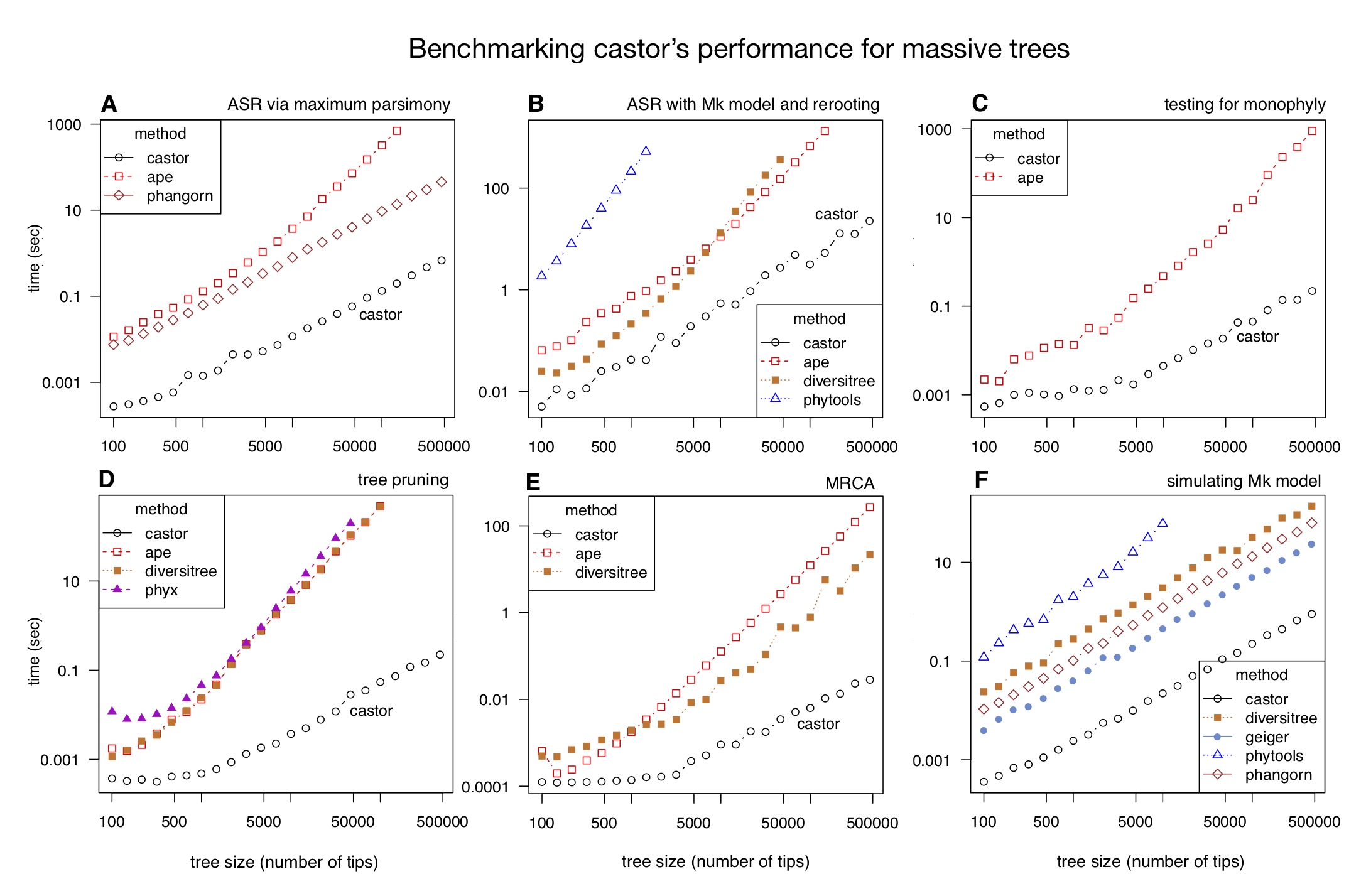
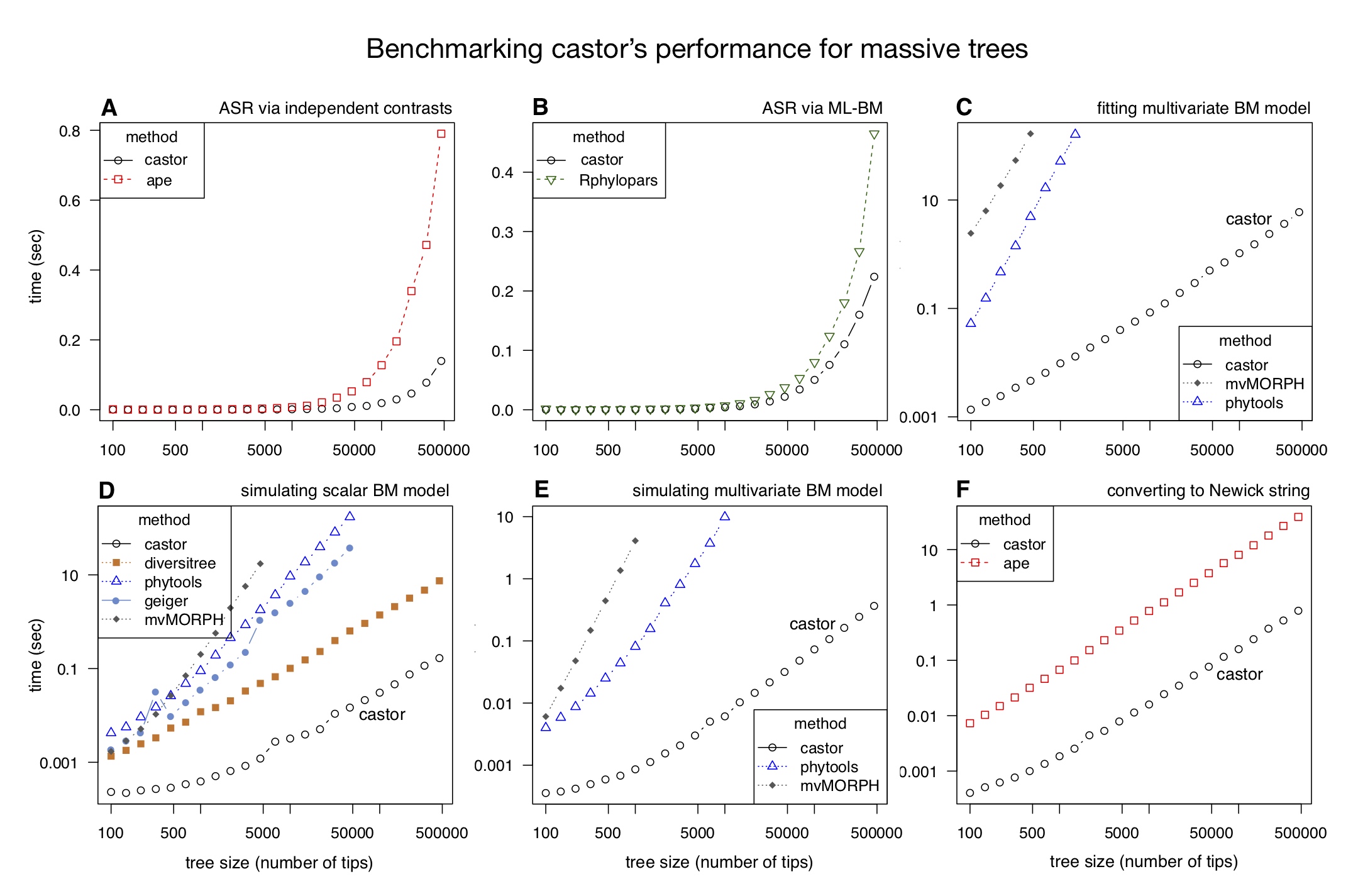
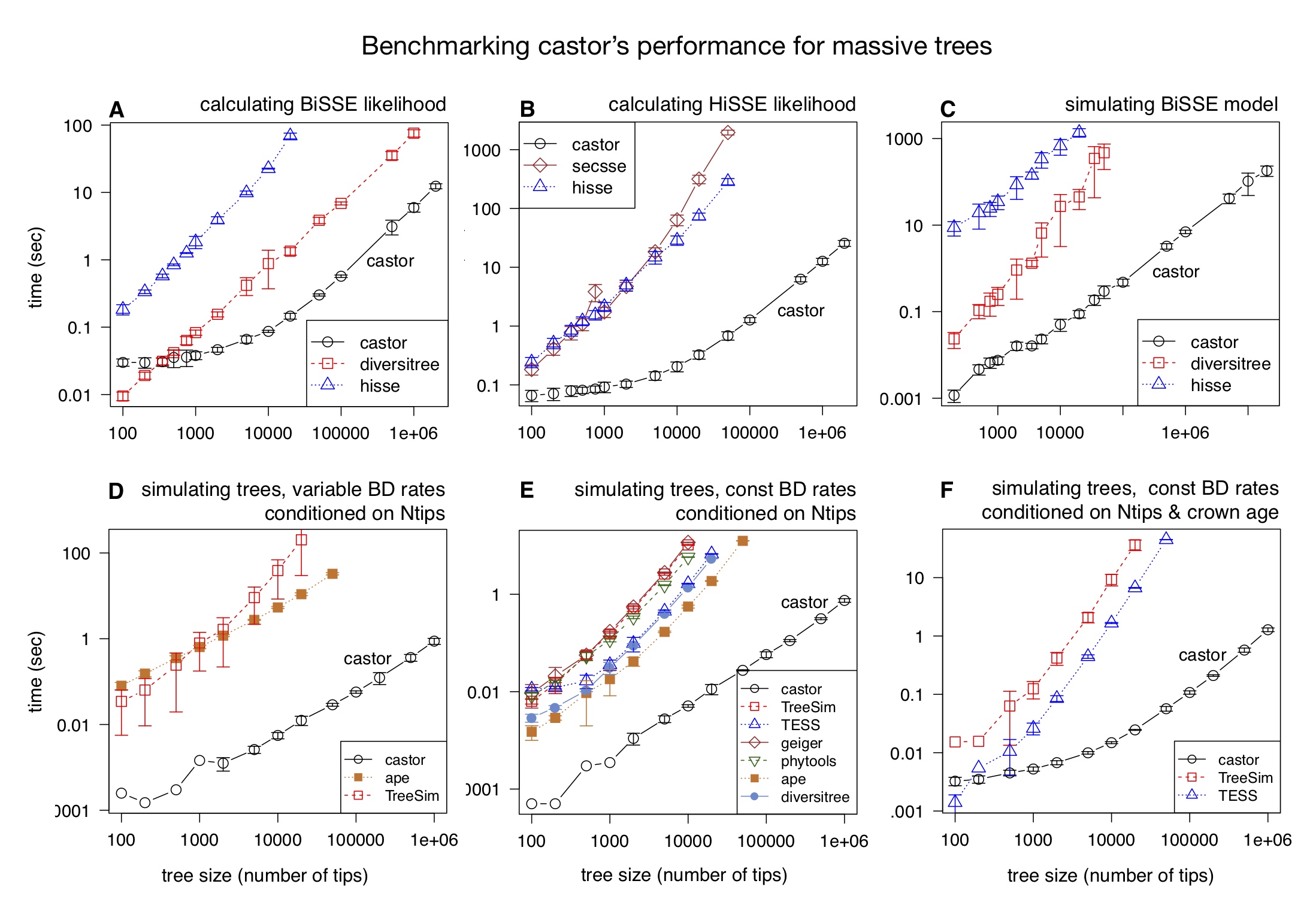
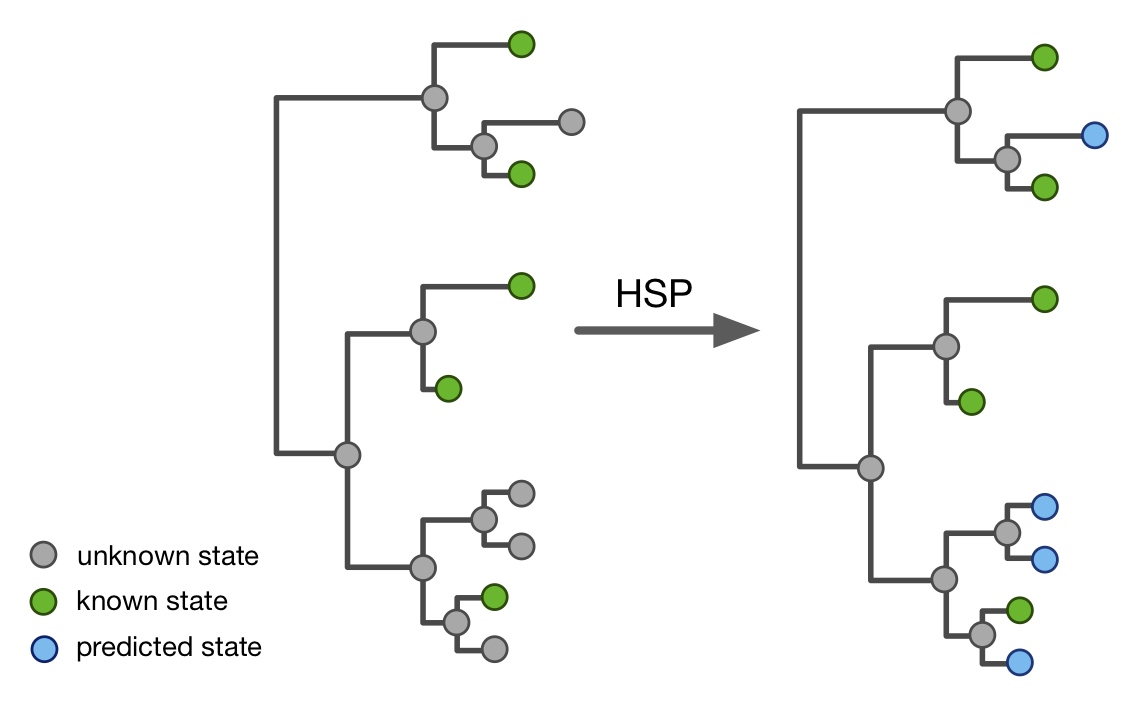
| castor |
|
|
 |
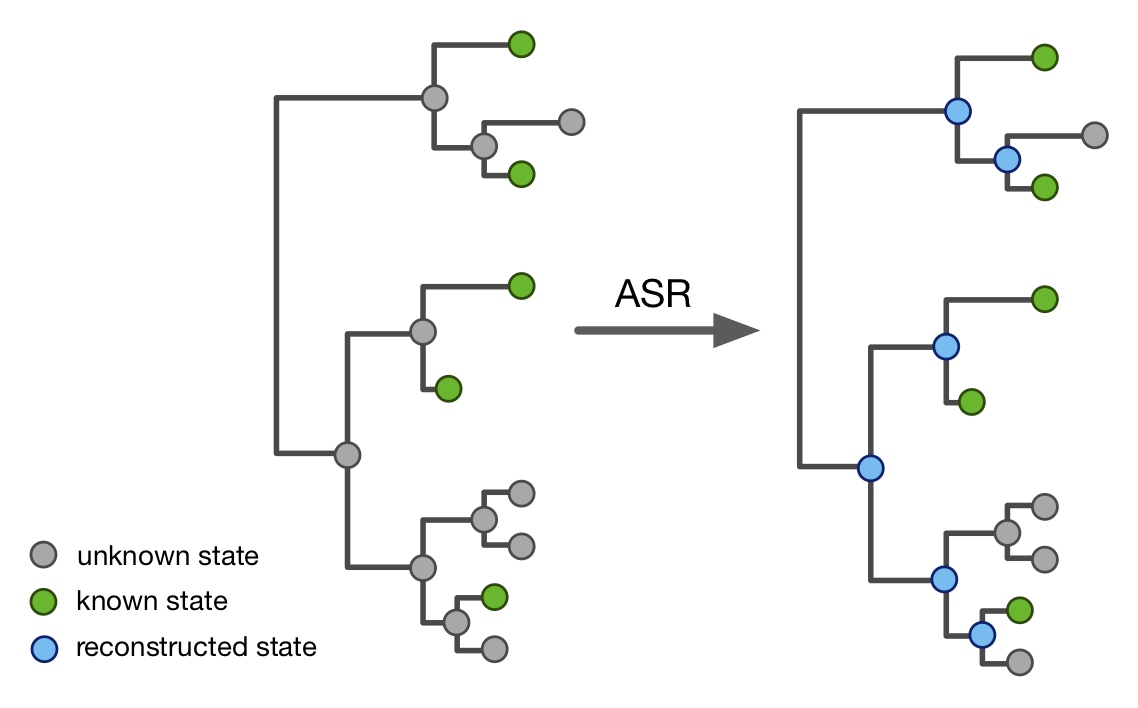
| An R package for efficient phylogenetics on large trees. |
|
| Reference: Louca, S., Doebeli, M. (2018). Efficient comparative phylogenetics on large trees. Bioinformatics 34:1053-1055 |
|
×
×
×
×
×
|
|
|
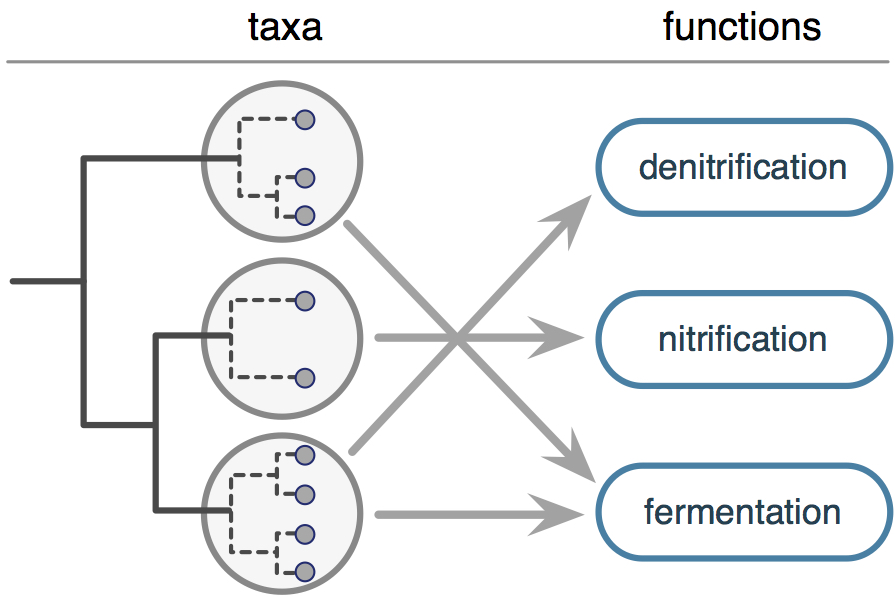
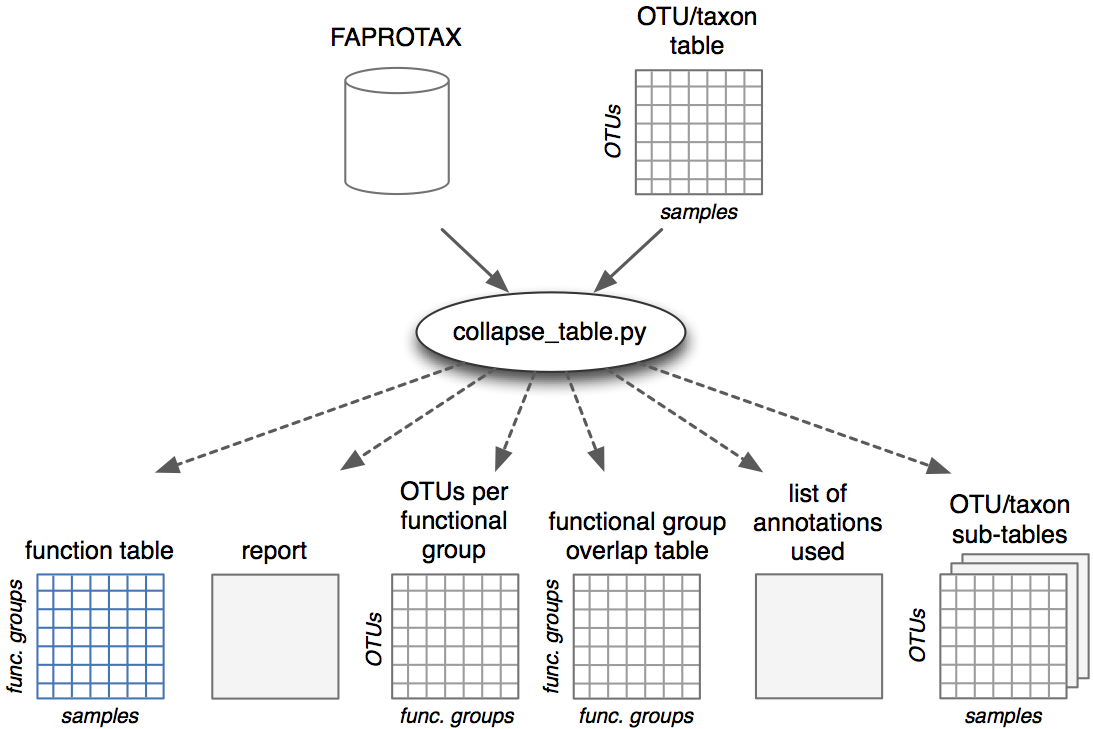
| FAPROTAX |
|
|
 |
| Database and software for mapping prokaryotic taxa to metabolic functional groups. |
|
| Reference: Louca, S., Parfrey, L. W., Doebeli, M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272-1277 |
|
×
×
|
|
|
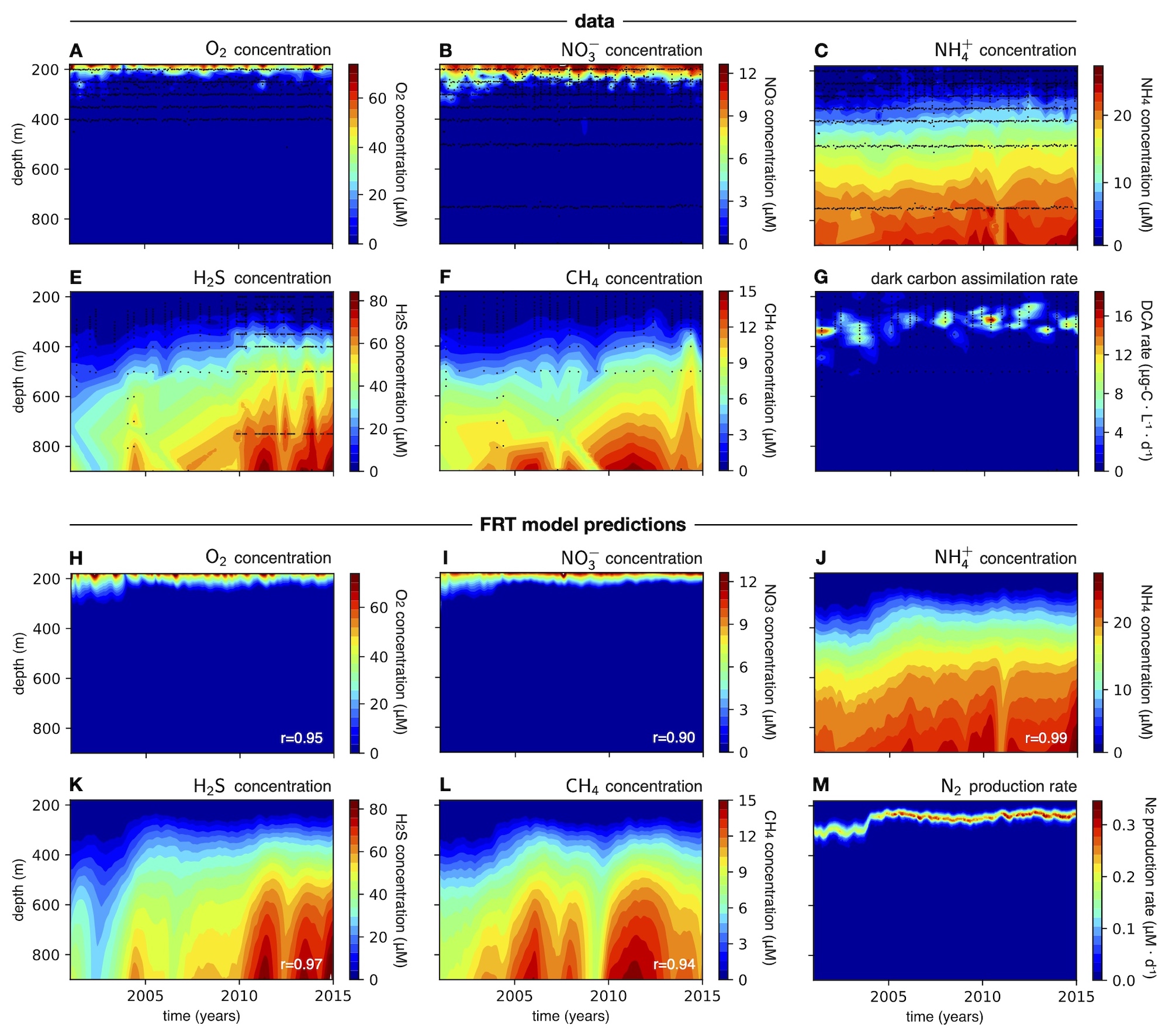
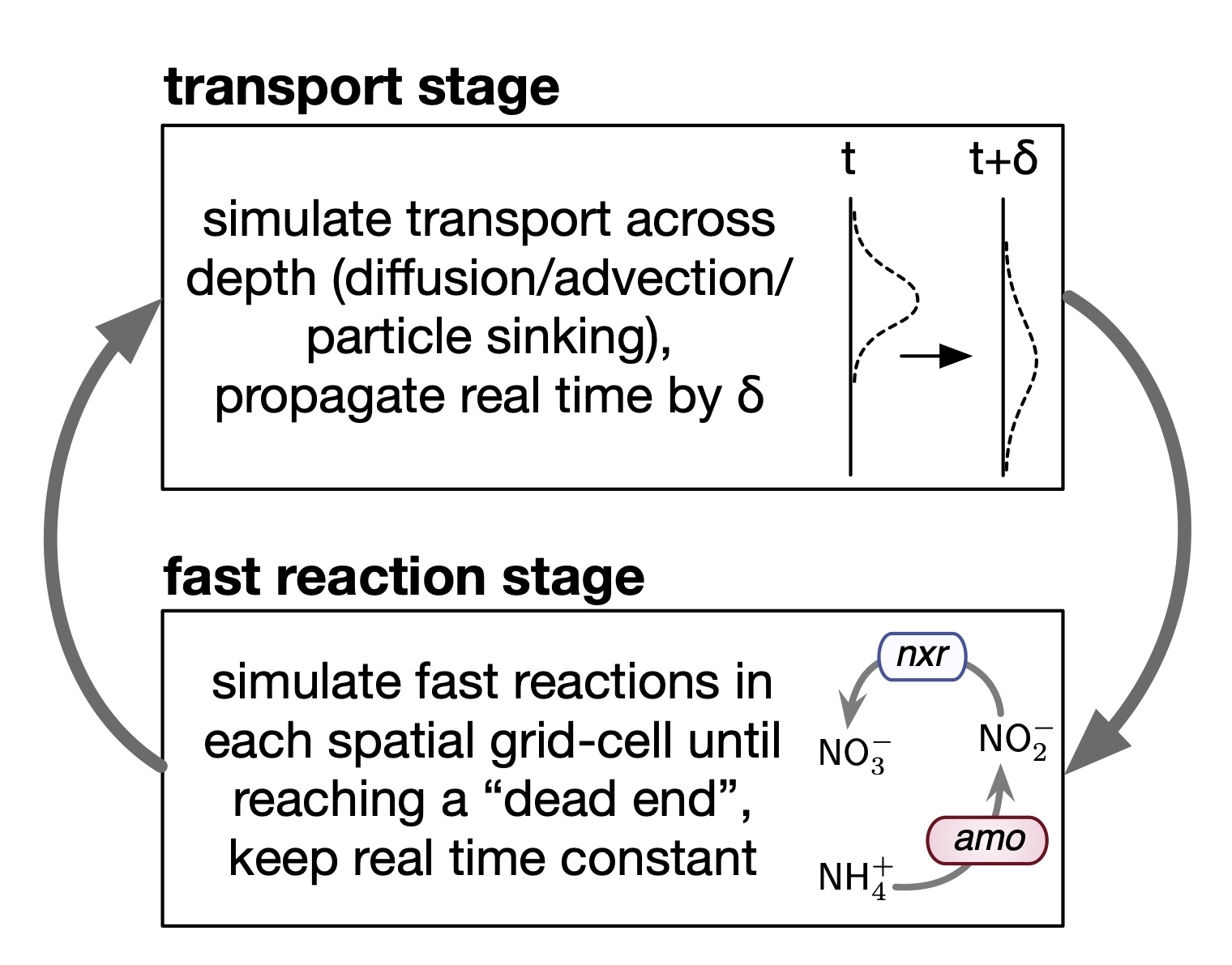
| Fast-reaction-transport modeling |
|
|
 |
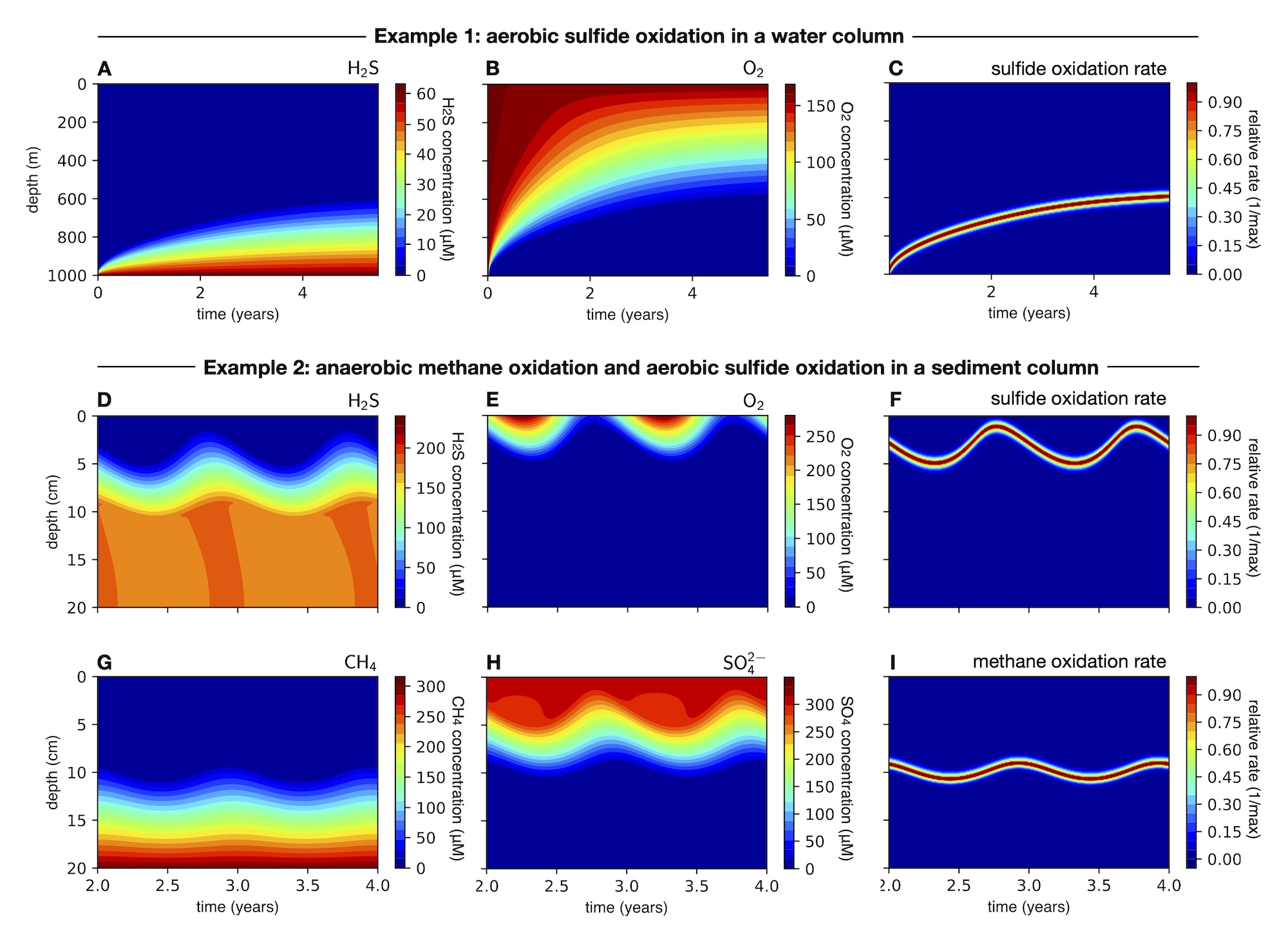
| MATLAB code demonstrating fast-reaction-transport biogeochemical models in 1-dimensional water and sediment columns. |
|
| Reference: Louca, S., Taylor, G. T, Astor, Y. M., Buck, K., Muller-Karger, F. E. (2022). Transport-limited reactions in microbial systems. Environmental Microbiology 25:268-282 |
|
×
×
×
|
|
|
| MCM (Microbial Community Modeler) |
|
|
 |
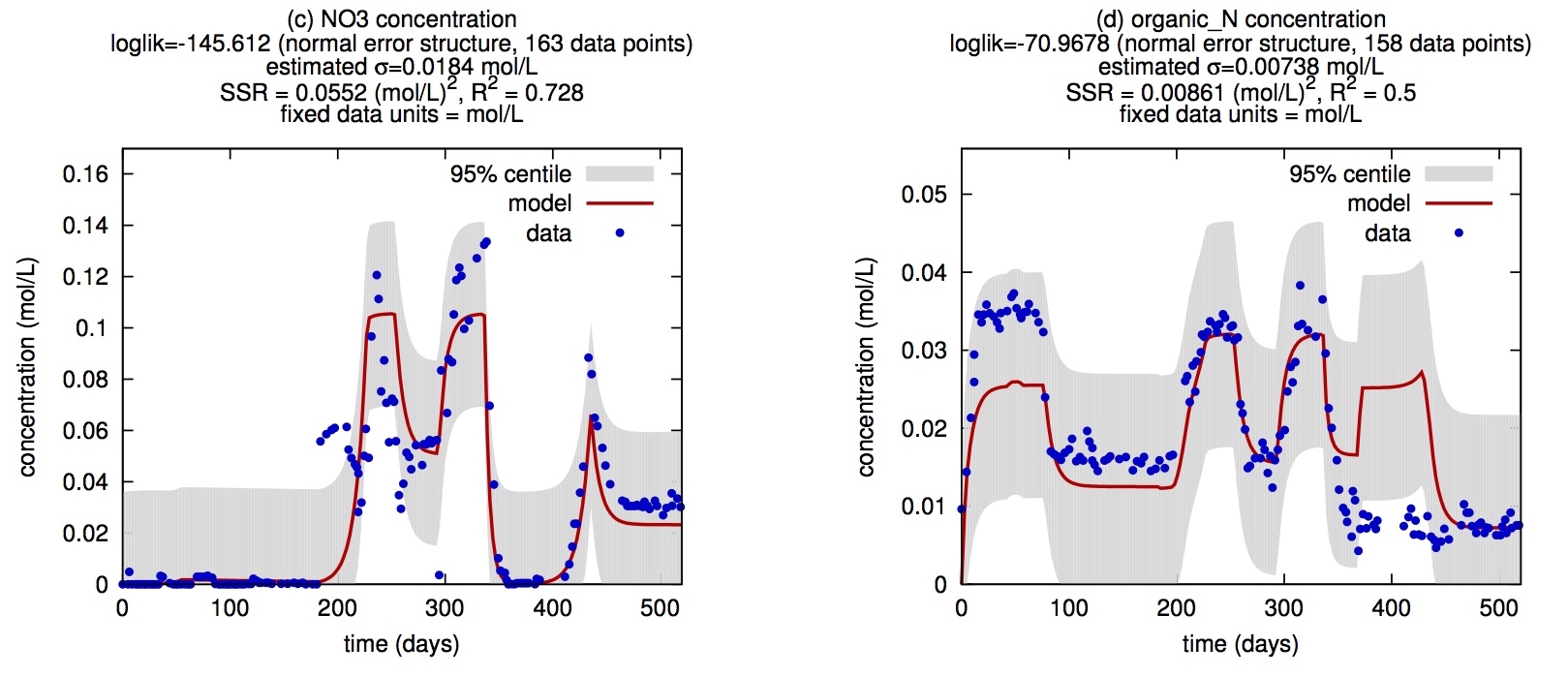
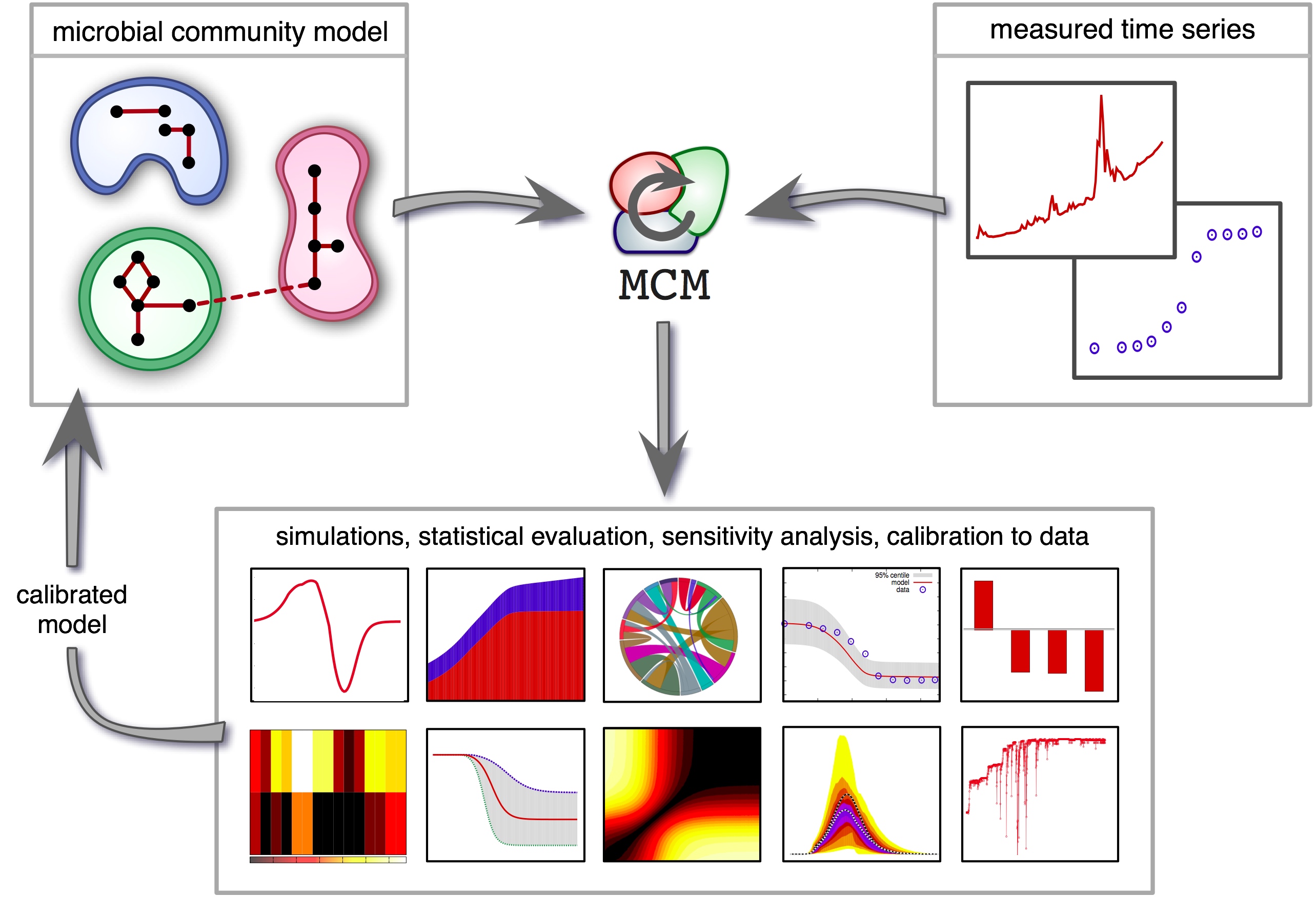
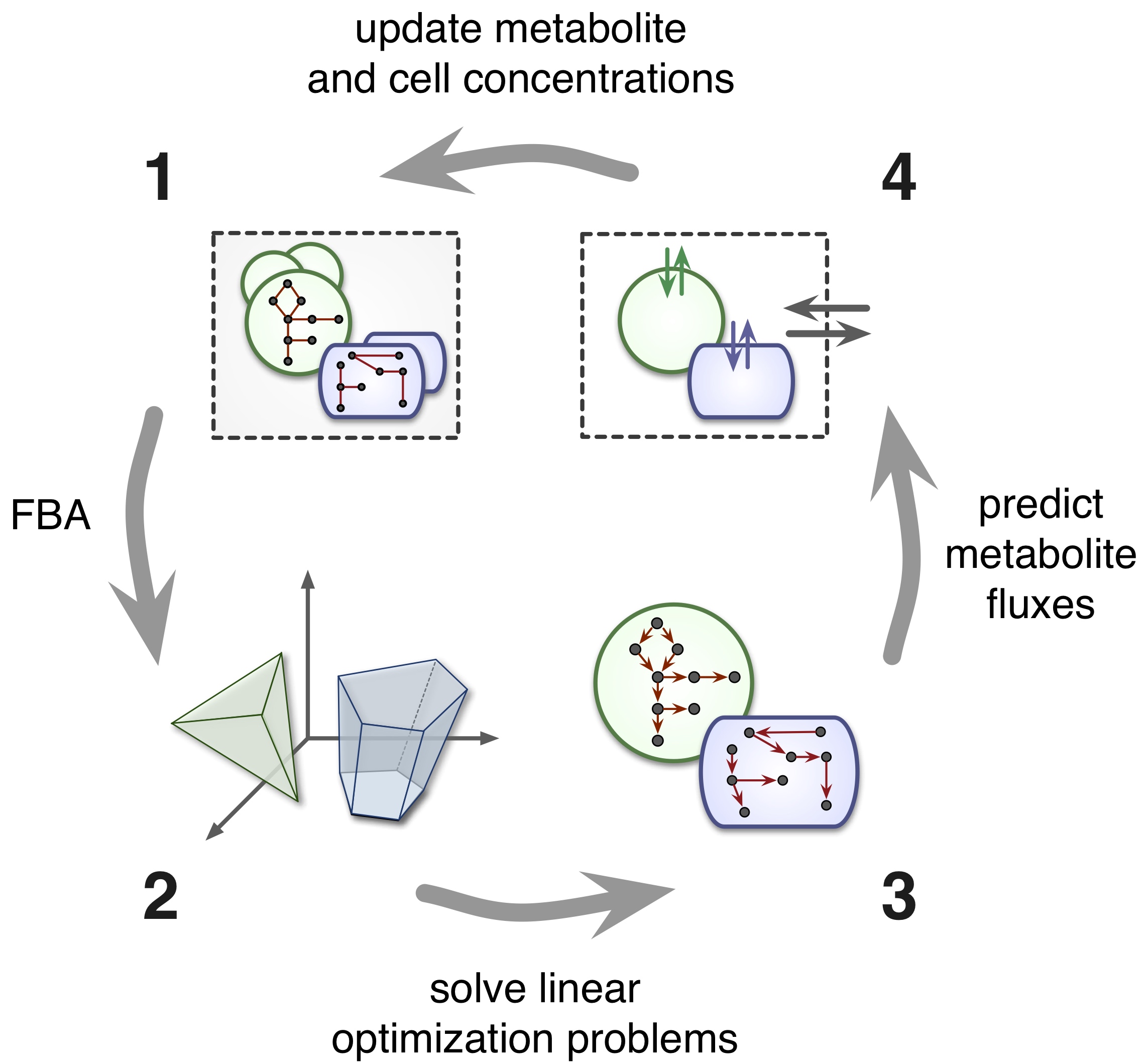
| A computational framework for modeling microbial communities using dynamic flux balance analysis of genome-based cell models, in the context of a dynamical environment. Classical functional group models are also supported. |
|
| Reference: Louca, S., Doebeli, M. (2015). Calibration and analysis of genome-based models for microbial ecology. eLife 4:e08208 |
|
×
×
×
|
|
|
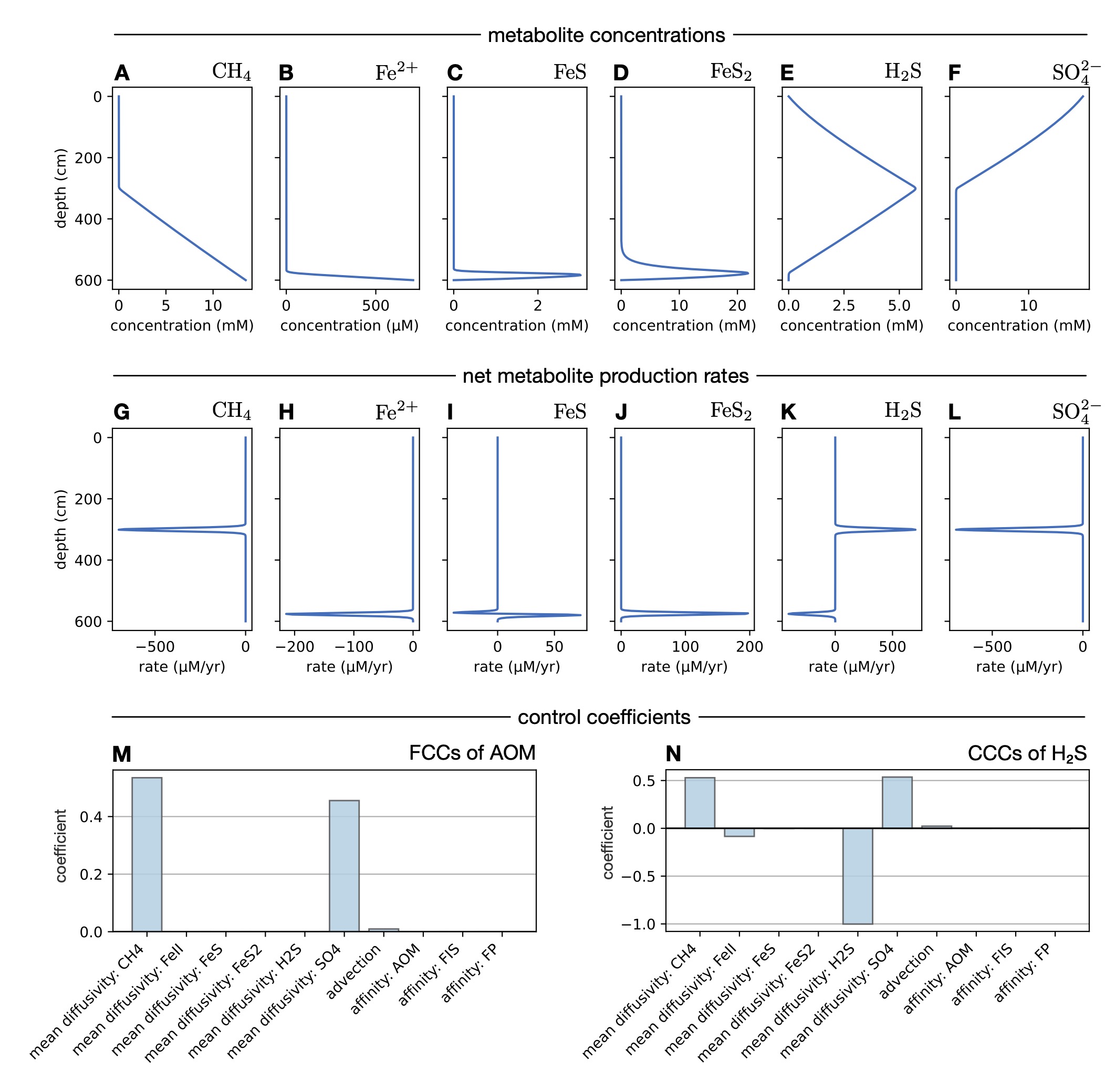
| Metabolic control analysis of biogeochemical systems |
|
|
 |
| Python code for simulating and performing metabolic control analysis of biogeochemical reaction-advection-diffusion models, at steady state. |
|
| Reference: Louca, S. (2025). Metabolic control analysis of biogeochemical systems. Communications Earth & Environment 6:161 |
|
×
×
|
|
|
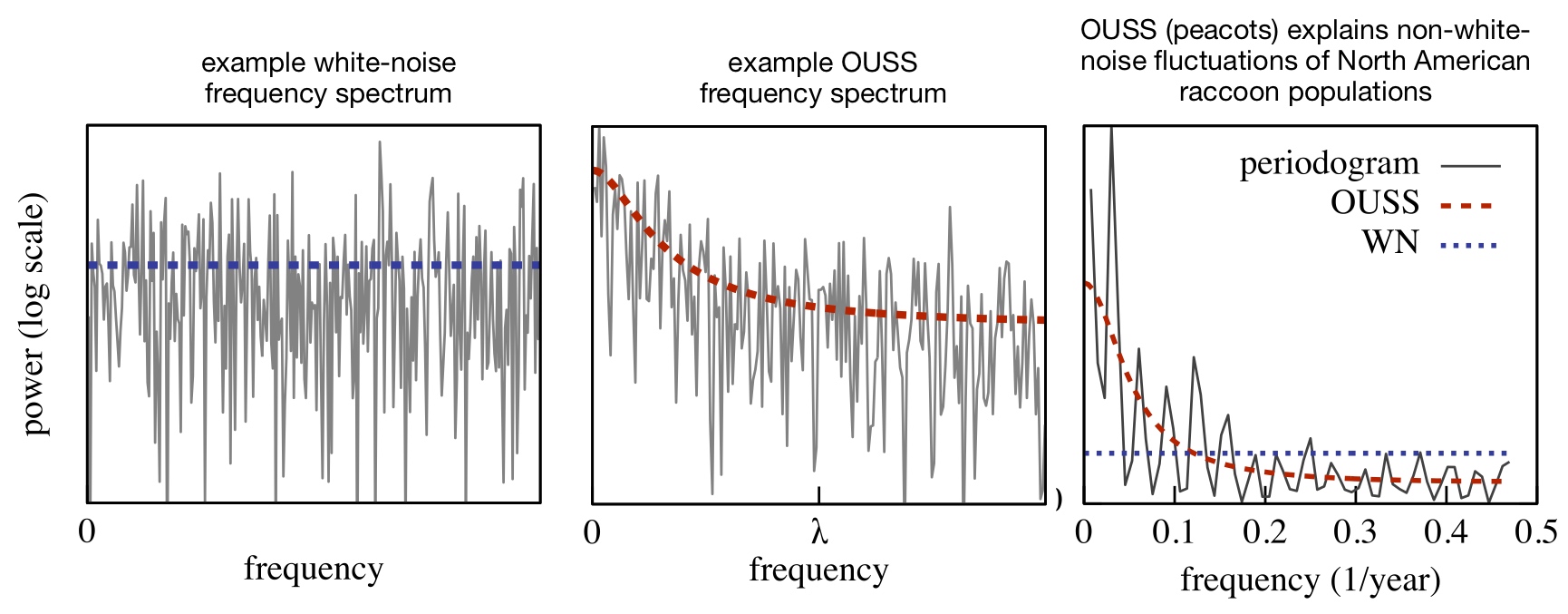
| peacots (Periodogram Peaks in Correlated Time Series) |
|
|
 |
| An R package for detecting cyclicity in time series using the Ornstein-Uhlenbeck state-space model (rather than simple white noise) as a null hypothesis. |
|
| Reference: Louca, S., Doebeli, M. (2015). Detecting cyclicity in ecological time series. Ecology 96:1724-1732 |
|
×
|
|
|
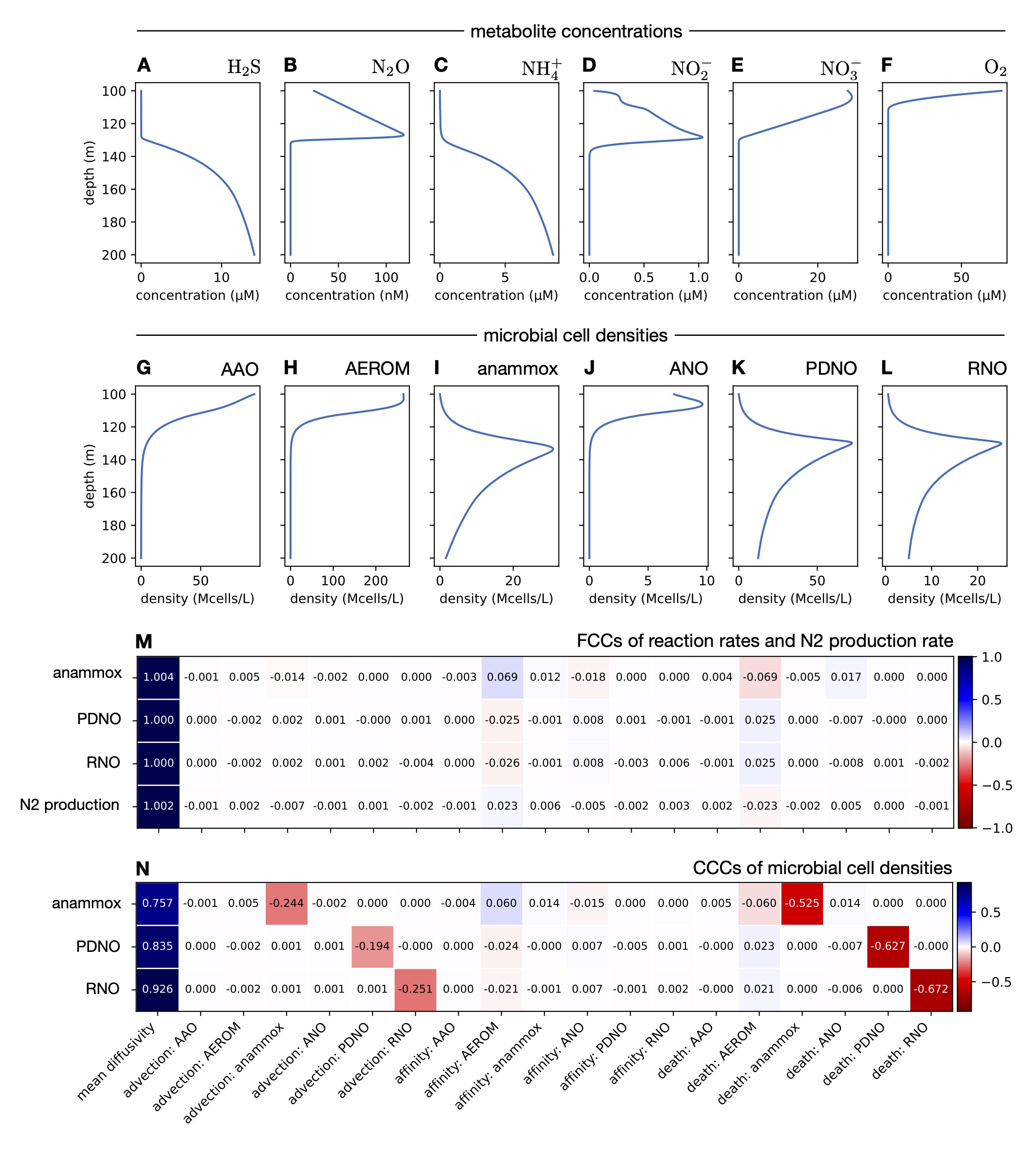
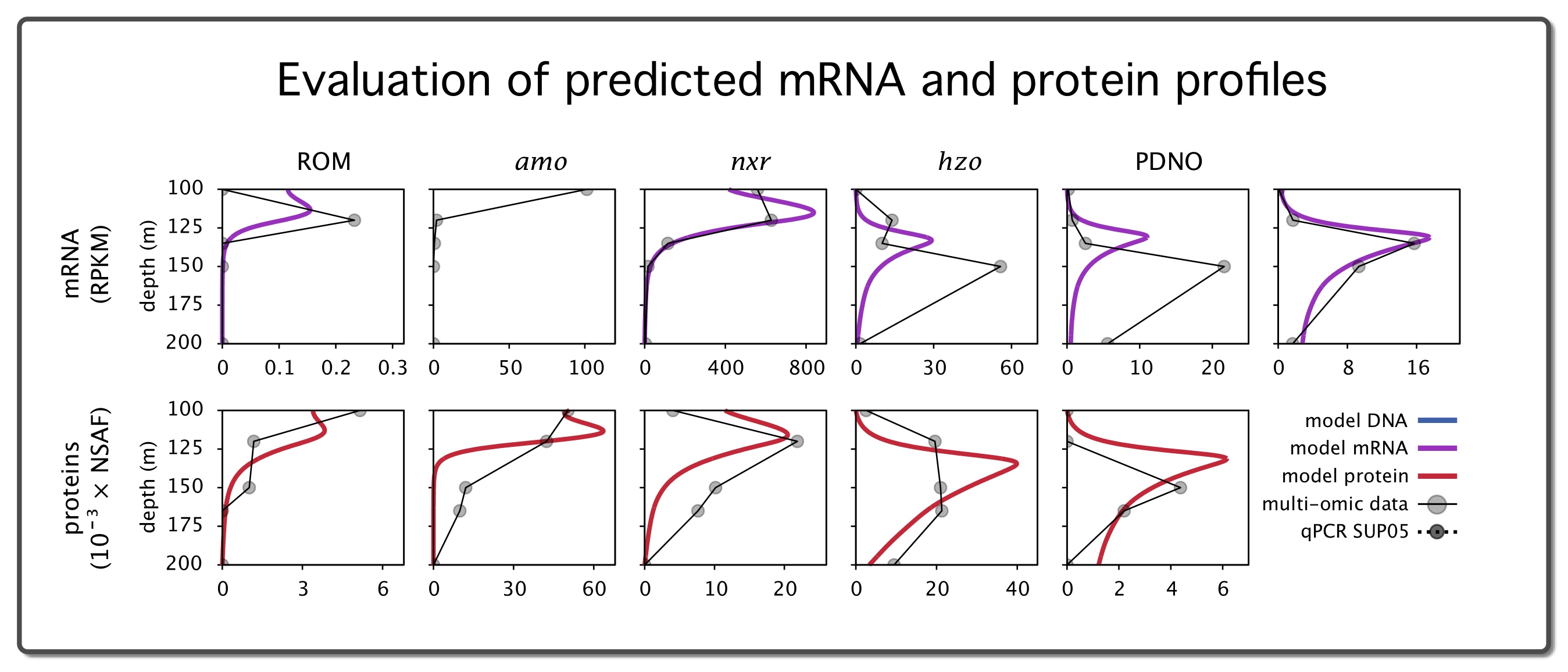
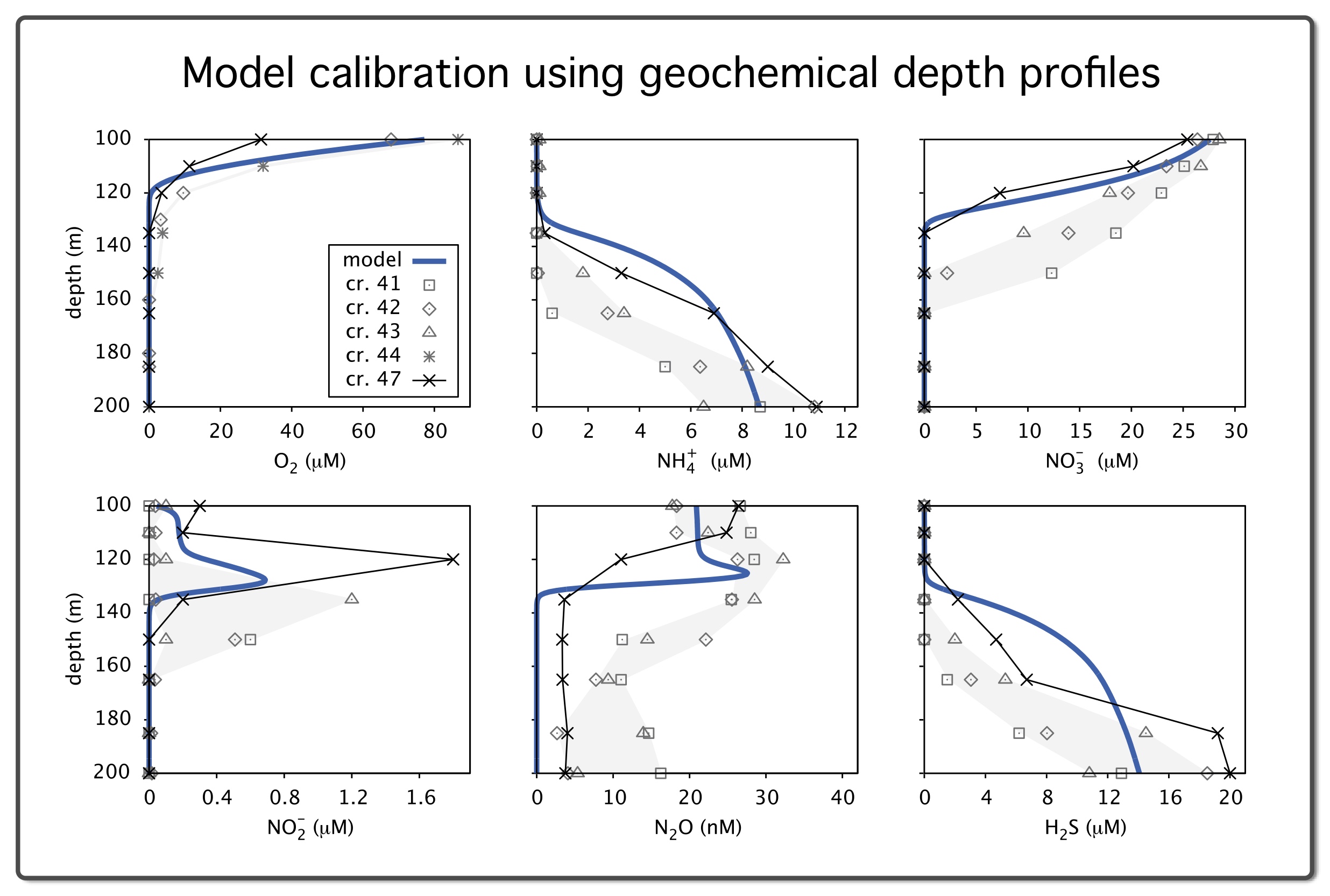
| Saanich GCM (Gene Centric Model) |
|
|
 |
| Gene-centric biogeochemical model (MATLAB code) for the Saanich Inlet seasonally anoxic water column, under steady state conditions. |
|
| Reference: Louca, S., Hawley, A. K., Katsev, S., Torres-Beltran, M., Bhatia, M. P., Kheirandish, S., Michiels, C. C., Capelle, D., Lavik, G., Doebeli, M., Crowe, S. A., Hallam, S. J (2016). Integrating biogeochemistry with multiomic sequence information in a model oxygen minimum zone. PNAS 113:E5925-E5933 |
|
×
×
|
|
|
|
|
| |
|
Louca lab. Department of Biology, University of Oregon, Eugene, USA
© 2025 Stilianos Louca all rights reserved
|
|
|



