|
|
| Cariaco Metabolic Modeling - MATLAB code |
This MATLAB code is provided as supplementary material to the following articles:
-
Louca, S., Astor, Y. M., Doebeli, M., Taylor, G. T., Scranton, M. I. (2019). Microbial metabolite fluxes in a model marine anoxic ecosystem. Geobiology 17:628-642
-
Louca, S., Scranton, M. I., Taylor, G. T., Astor, Y. M., Crowe, S. A., Doebeli, M. (2019). Circumventing kinetics in biogeochemical modeling. PNAS 116:11329-11338
The main implemented workflows are:
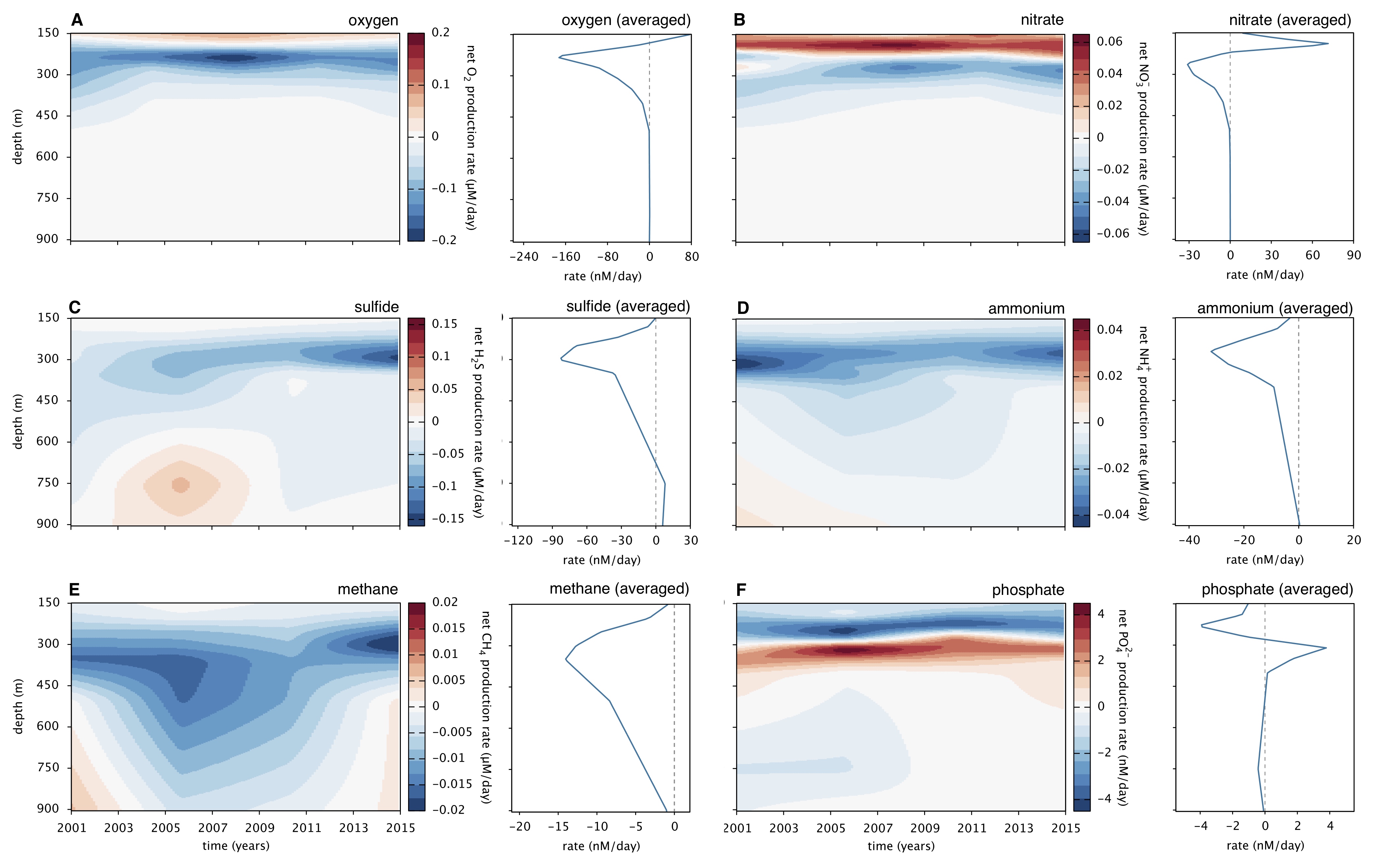
- ILTM: Inverse Linear Transport Modeling, for reconstructing net production rates of compounds diffusing/advecting in 1-dimensional systems, based on concentration measurements at discrete spacetime points.
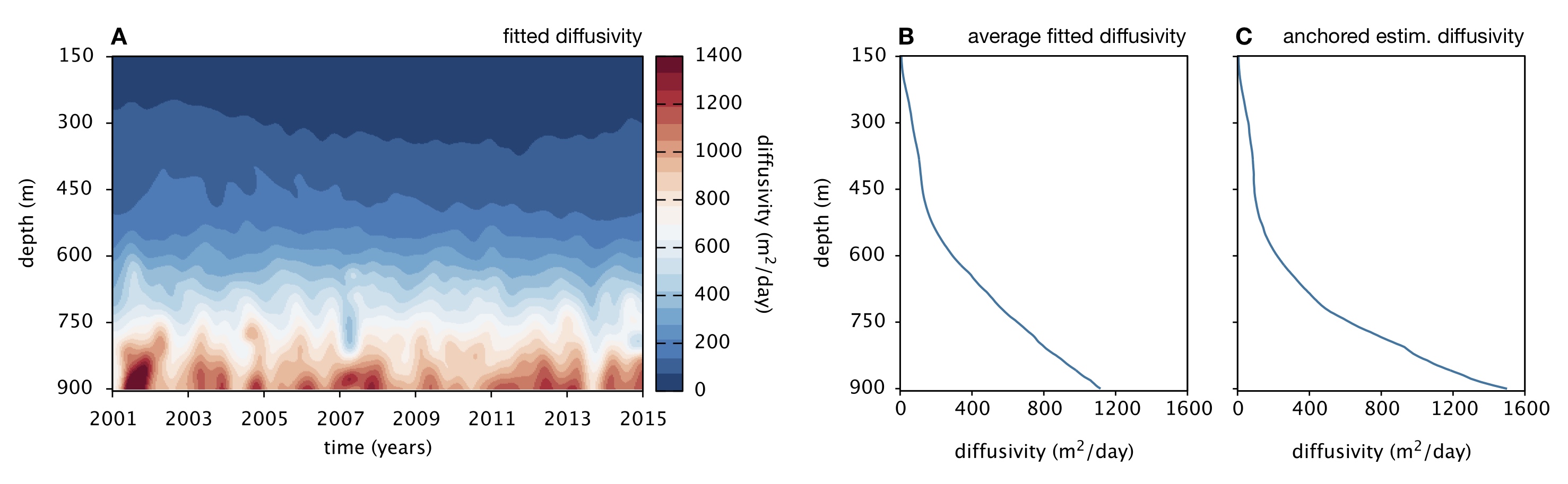
- Estimating diffusion coefficients across space and time based on measurements of some conserved tracer (such as salinity).
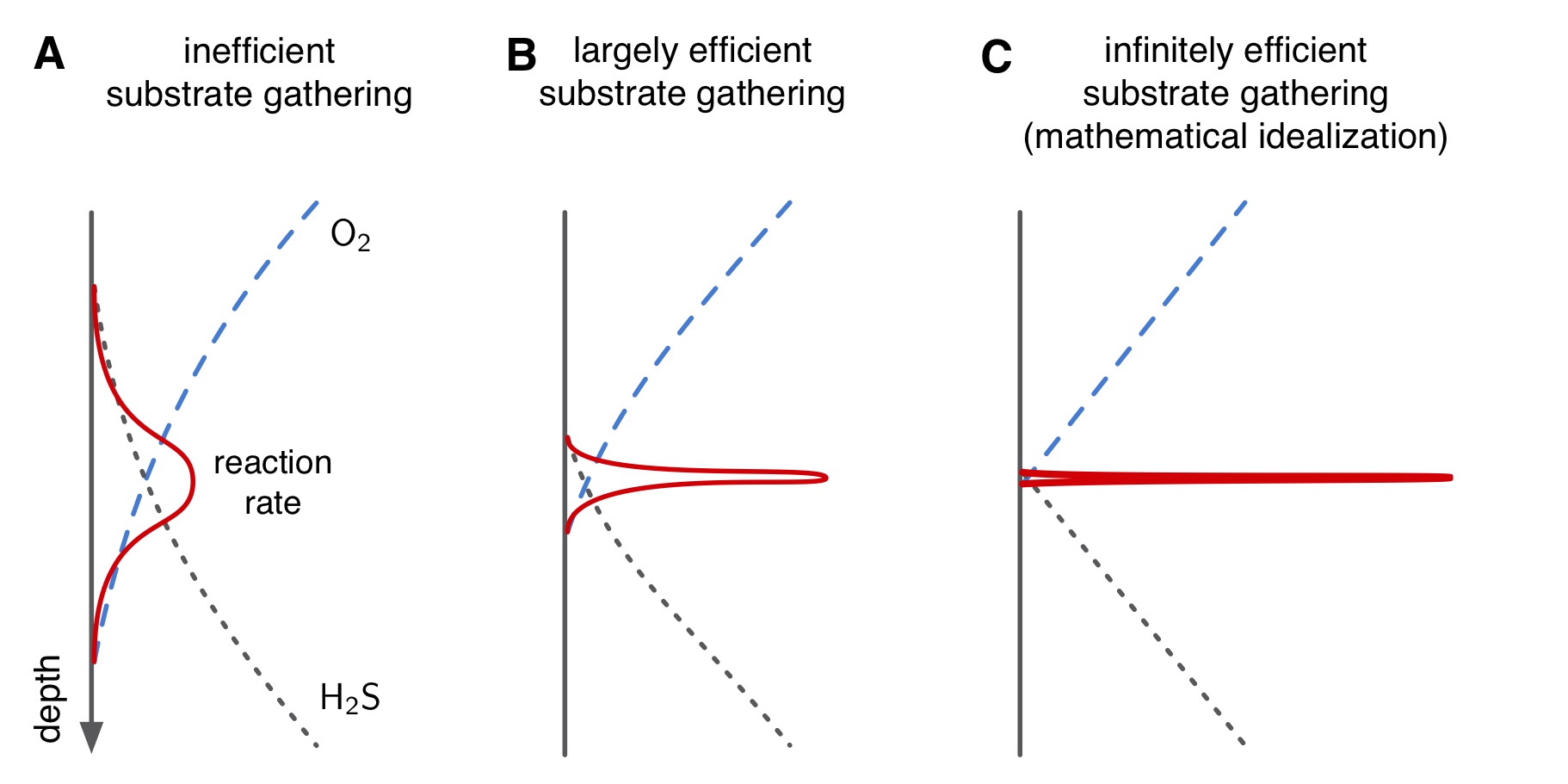
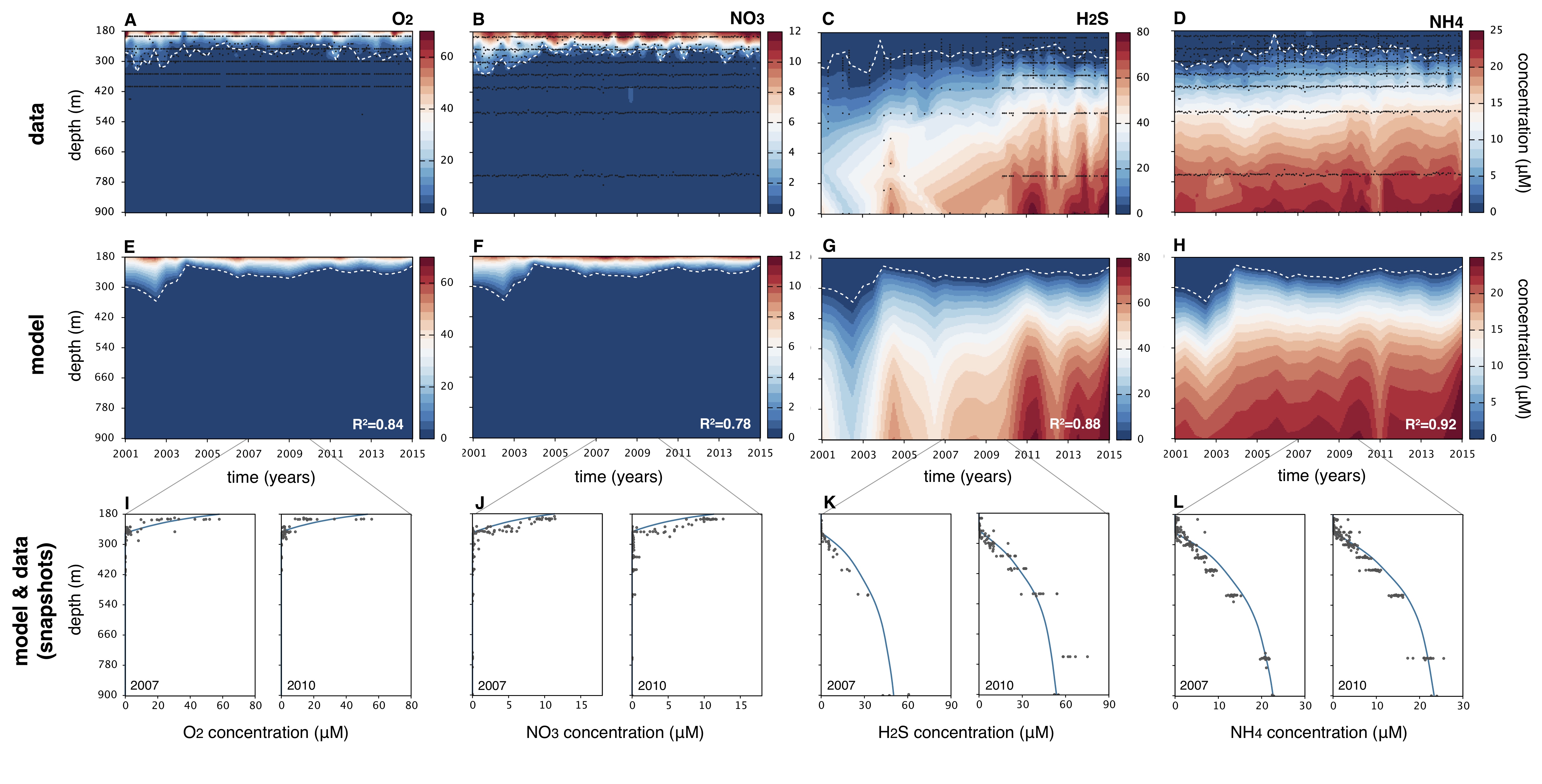
- SMF: Spatial Metabolic Flux modeling of 1-dimensional microbial systems with diffusive/advective metabolite transport near steady state. This modeling framework predicts microbially driven metabolic fluxes across space, solely based on chemical boundary conditions at the top and bottom, but without knowledge of microbial kinetics, and without any parameter fitting. The key assumption of the SMF framework is that metabolic reactions occur at discrete points ("hotspots"), outside of which each reaction is limited by at least one substrate. By assuming that microbial populations are infinitely efficient at gathering substrates, reaction- and bio-kinetics are essentially "shortcutted".
The above workflows are implemented as largely independent modules. All workflows are exemplified using physical and chemical data from the Cariaco Time Series Program, but can be adjusted to other similar 1-dimensional systems, such as meromictic lakes or marine sediments. The code can account for variation of the lateral (cross-sectional) basin/lake area with depth.
The SMF workflow, implemented in the MATLAB file fitSMFmodel.m, can handle an arbitrary number of metabolites, reactions and hotspots, separate diffusion coefficients and advection speeds for each metabolite (each of which can vary with depth), optional constraints on hotspot locations and/or individual reaction rates, various combinations of boundary conditions (Dirichlet and/or Robin) and supports various types of stress functions. The code can optionally take into account available depth-profile data to improve predictive accuracy.
A regular MATLAB installation should be sufficient to run this code.
The code has been tested in MATLAB R2015b, on Mac OS 10.13.6.
Other MATLAB versions may require some slight modifications of the code.
Please consult the README files included in the downloads below for technical details and license agreements.
Downloads:
 | Complete MATLAB code, including input data for ILTM and SMF analyses in Cariaco. |
 | Code demonstrating SMF for a simple hypothetical scenario (one reaction). |
 | Code demonstrating SMF for a slightly more complex scenario (two coupled reactions, two hotspots). |
 | Code demonstrating SMF for two coupled reactions and two hotspots, and an explicitly added methane influx due to remineralization of organic matter. |
 | Code demonstrating SMF for indirect oxidation of H2S with O2 via a Mn redox shuttle (two reactions, two hotspots, MnO2 transport is purely advective). |
|
|
 ×
 ×
|
|
|
Louca lab. Department of Biology, University of Oregon, Eugene, USA
© 2025 Stilianos Louca all rights reserved
|
|
|



