|
|
| Metabolic control analysis of biogeochemical systems - Python code |
This python code is provided as supplementary material to the following article:
Abstract:
Many biogeochemical systems, including the ocean carbon and nitrogen cycles, involve multiple biological and abiotic processes operating at different scales, such as hydrodynamic transport and molecular diffusion across space, abiotic chemical transformations, as well as microbial metabolism and population dynamics.
Determining the influence of these processes on emergent system behavior is critical for model construction and for prioritizing efforts to determine physical/biological parameters.
Metabolic control analysis (MCA) is a well-established framework for quantifying the role of individual enzymes in cellular metabolic networks at steady state, but its applicability to biogeochemical systems has not been explored.
Here I show how the core concepts and theorems of MCA can be generalized to complex biogeochemical systems at arbitrarily large spatial scales, enabling insight into the roles of physical transport, microbial dynamics and chemical kinetics at organismal to planetary scales.
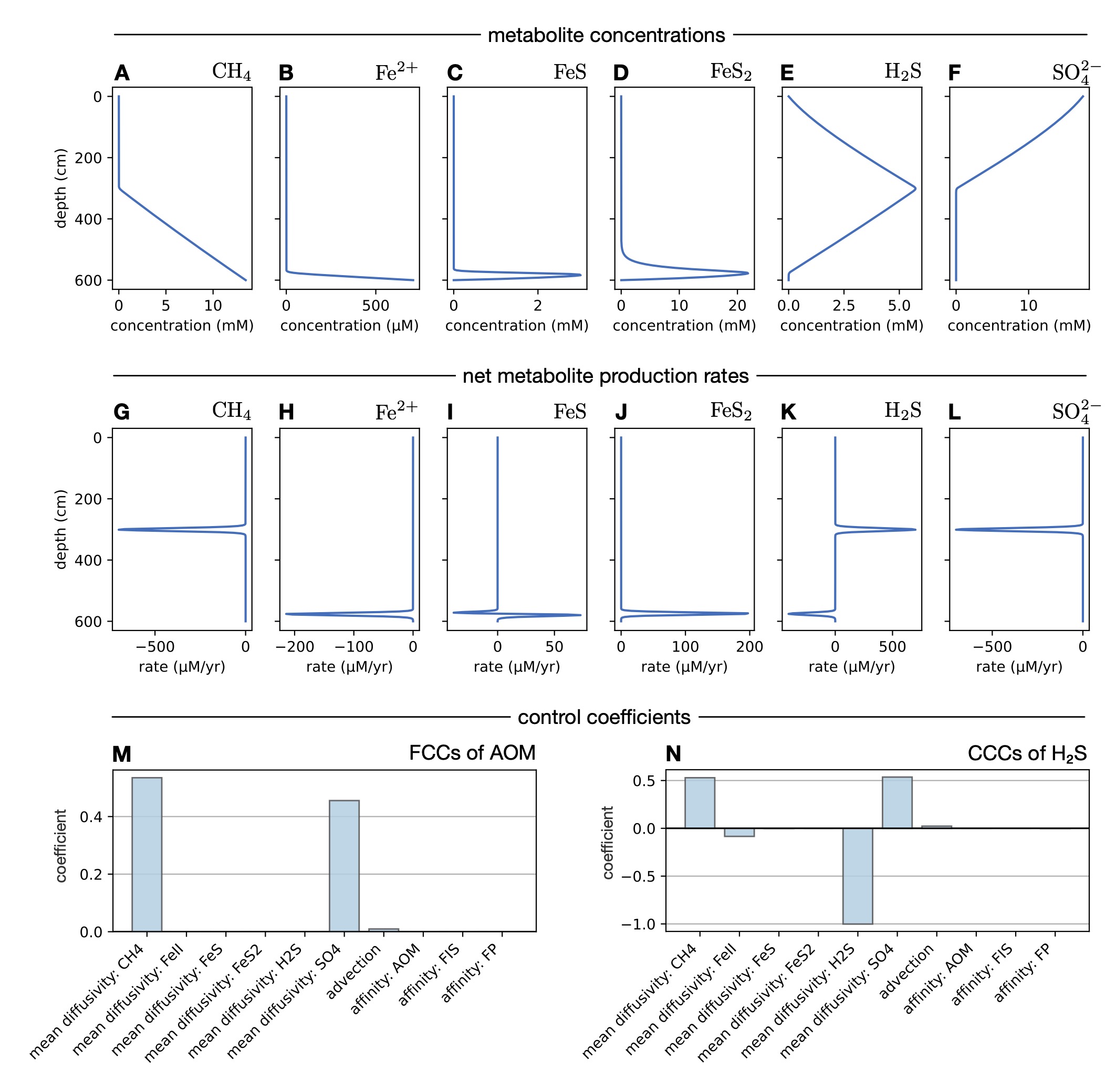
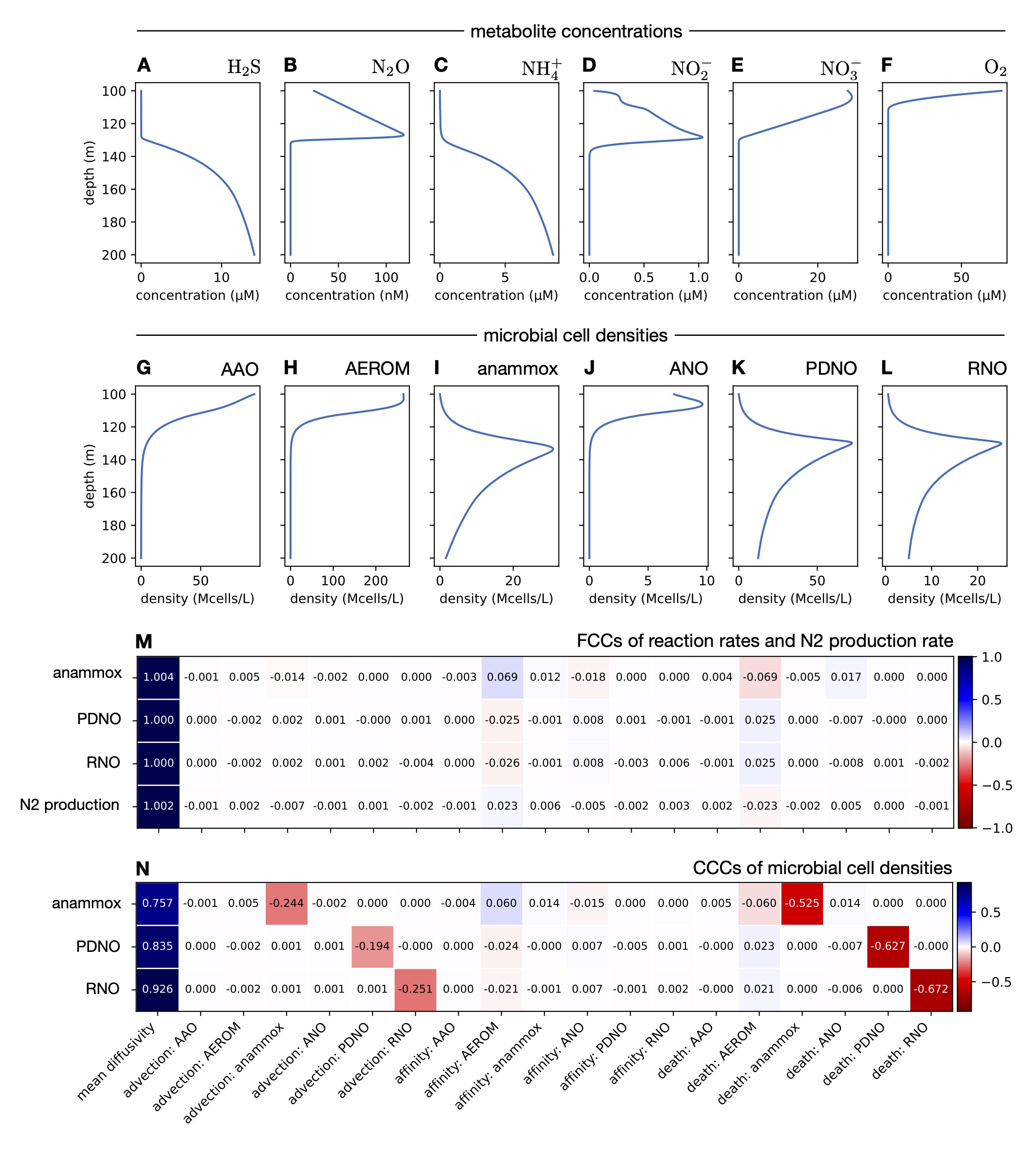
I demonstrate the power of this generalized MCA on two systems of major relevance to ocean biogeochemistry and climate research: sulfate methane transition zones in Black Sea sediments and a denitrifying oxygen minimum zone in Saanich Inlet.
MCA reveals that physical transport is by far the greatest rate-limiting factor for sulfate-driven methane oxidation in the first system and for fixed nitrogen loss in the second system.
The code provided below is used to simulate the models described in the above article to steady state, and perform metabolic control analysis.
The code only requires a standard python installation with common packages such as numpy, scipy and matplotlib. The code has been tested in python 3.11, MacOS 15.4.
For further usage instructions as well as the license agreement see the header of the workflow.py code file.
Downloads:
 | Complete python code. |
|
|
 ×
 ×
|
|
|
Louca lab. Department of Biology, University of Oregon, Eugene, USA
© 2025 Stilianos Louca all rights reserved
|
|
|



