|
|
| Castor - Efficient phylogenetics on large trees |
Castor is an R package for performing efficient phylogenetic analyses on massive trees, including up to millions of tips. Some of castor's functionalities are:
- Pruning tips, rerooting, collapsing closely related tips.
- Finding most recent common ancestors.
- Calculating distances from the tree root and pairwise tip distances.
- Calculating phylogenetic signal and mean trait depth (trait conservatism).
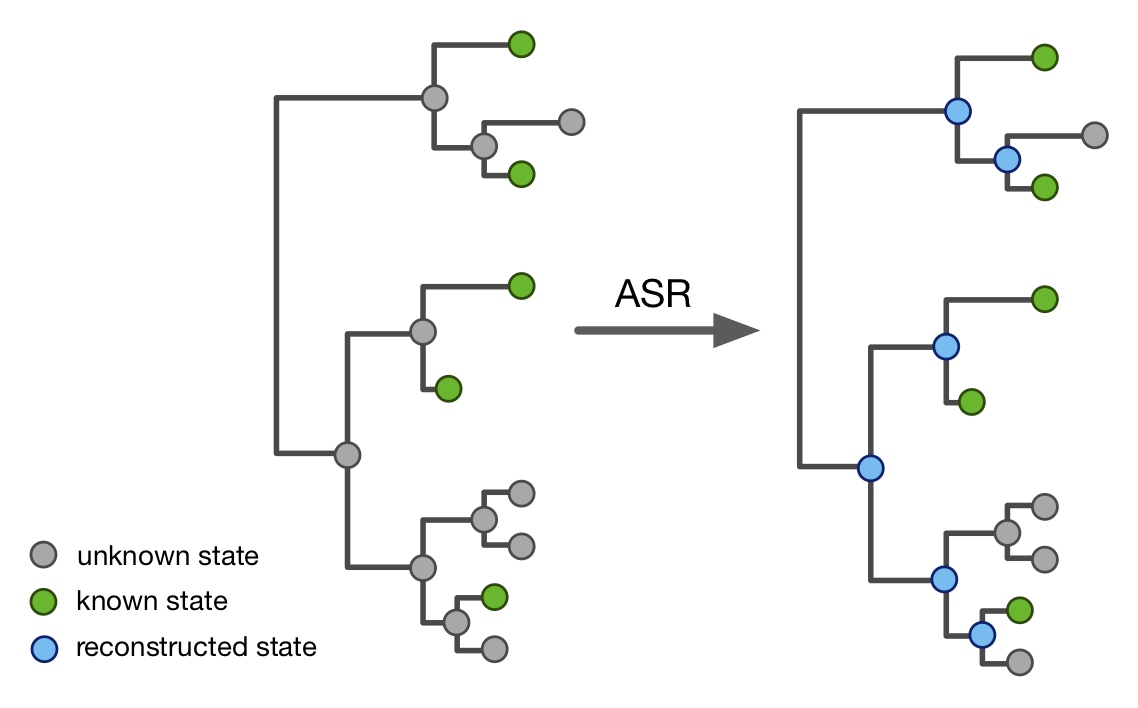
- Ancestral state reconstruction (ASR) of discrete and continuous traits.
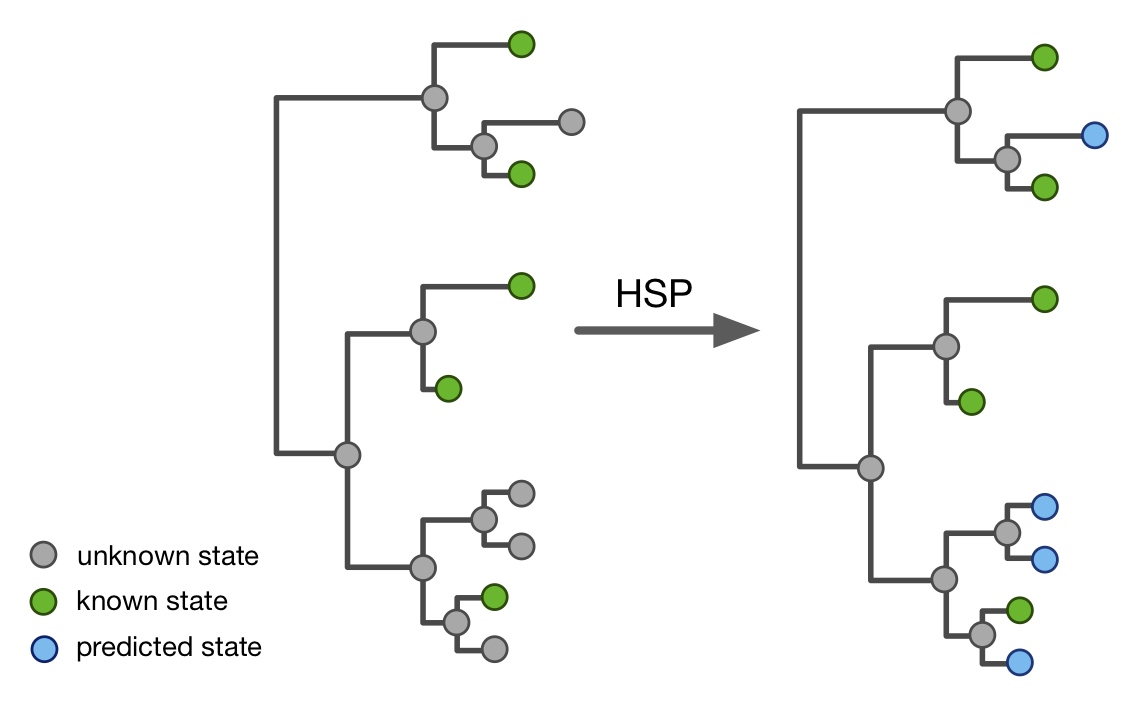
- Hidden state prediction (HSP) of discrete and continuous traits on tips.
- Simulating and fitting models of trait evolution.
- Simulating and fitting birth-death models (macroevolution)
- Simulating and fitting birth-death-sampling models (epidemiology).
- Generating random trees using birth-death models.
- Comparing trees, finding congruencies between trees (e.g. for dating).
- Writing and loading trees from files (Newick format).
- Fitting diffusion models of geographic dispersal to phylogeographic data.
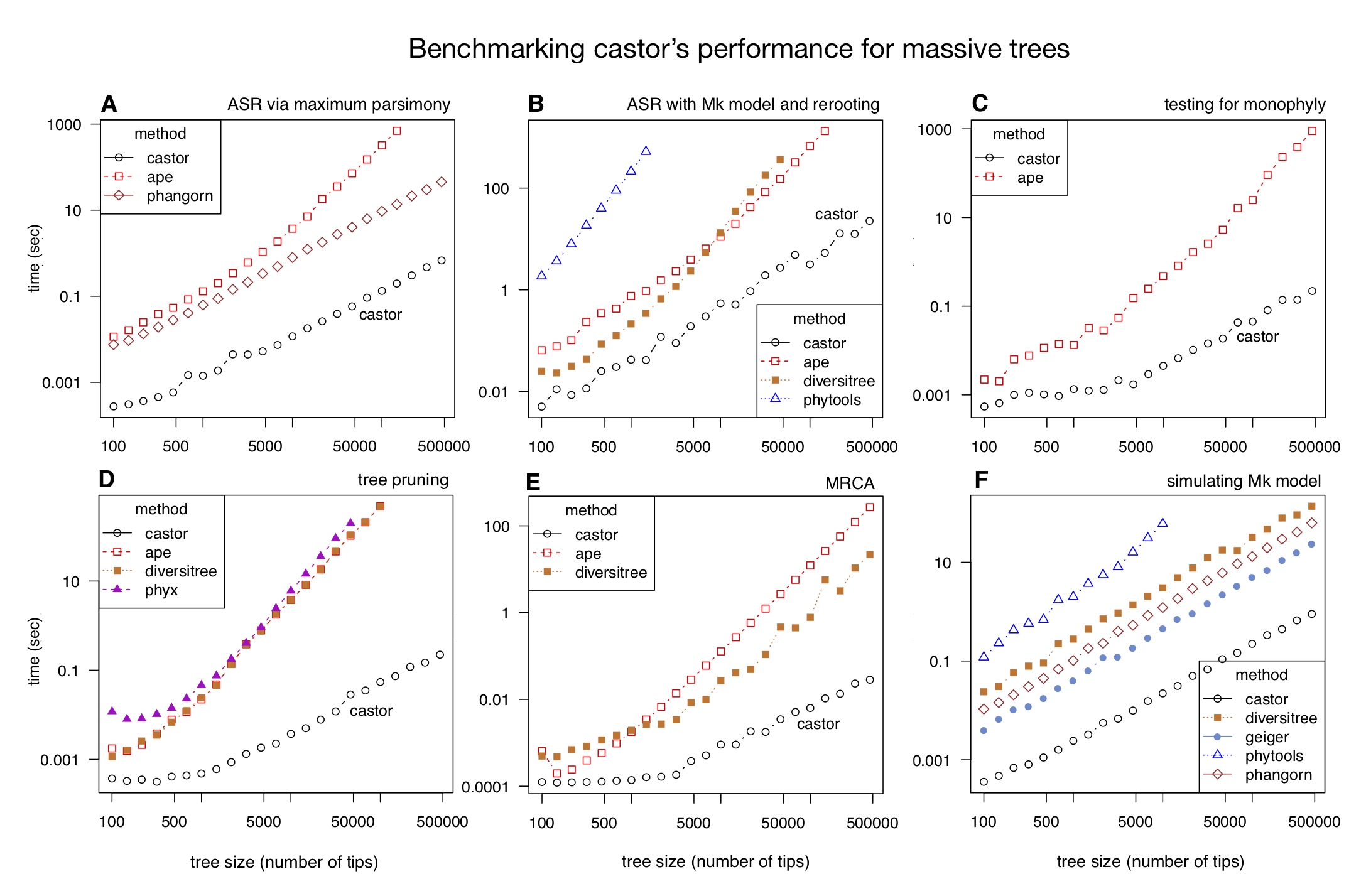
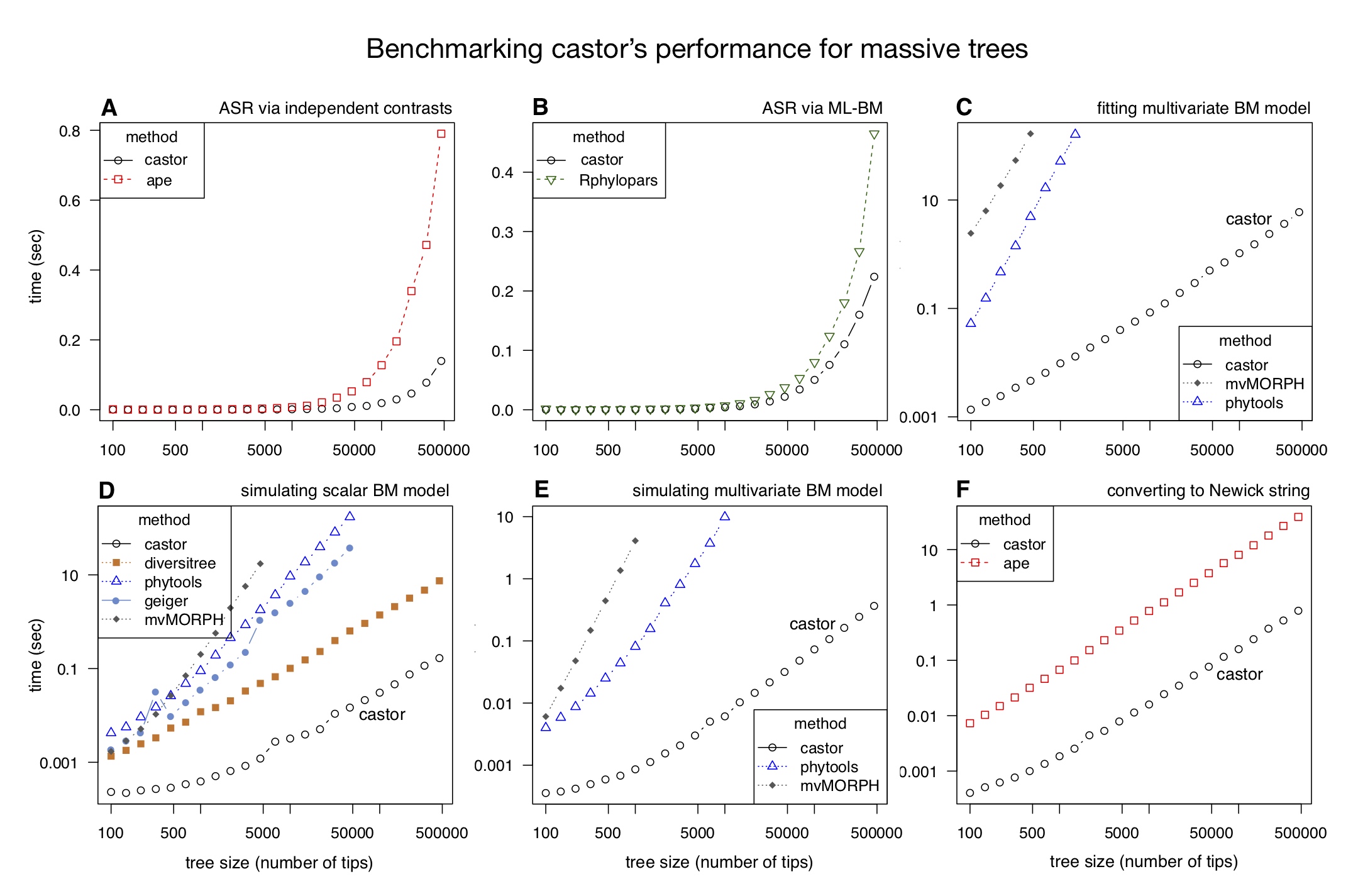
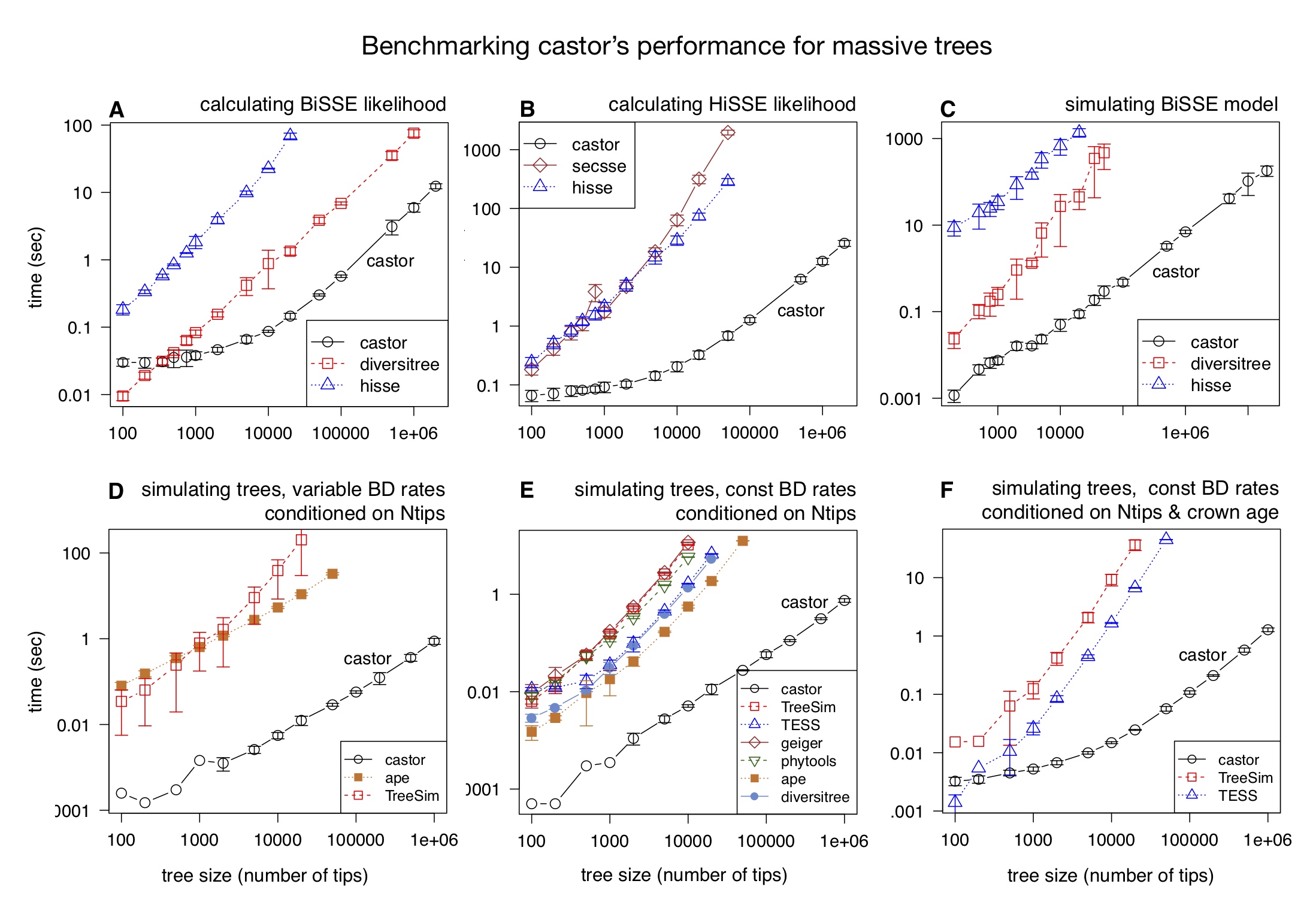
Most of castor’s functions exhibit a time complexity that scales linearly with tree size, in many cases being 100 to 1000 times faster than other comparable tools. For example, removing half of the tips from the SILVA tree (~500,000 tips) takes less than 1 sec on a modern laptop using castor and over 4 hours using other comparable R packages (as of 2018).
How can castor be so efficient?
Castor’s high efficiency is achieved in multiple ways.
First, dynamic programming algorithms are used wherever possible.
Second, most algorithms benefit from auxiliary internal data structures that are temporarily created on demand. While this sometimes adds a minor fixed cost, for larger trees this can dramatically reduce the scaling of computation time.
Third, in certain ASR algorithms involving rerooting (e.g. maximum-likelihood Mk models) redundant calculations are avoided by storing previously computed intermediate quantities.
Fourth, ASR of discrete traits using Mk models, which requires repeated exponentiation of the Markov transition matrix along each edge, was accelerated through an ad-hoc exponentiation algorithm that becomes highly efficient when the same matrix is exponentiated several times.
Fifth, castor is almost entirely implemented in C++, a programming language suitable for high-performance computations.
Citations:
Castor is described in the following papers:
Download:
Stable releases of castor are available on CRAN, and can be installed using the R command:
|
install.packages("castor", repos="http://cran.r-project.org")
|
Note that the latest version of castor may not yet be on CRAN.
The latest castor version (potentially beta) can be downloaded as a self-contained installation file here.
To load castor from the installation file into a single R session use:
|
devtools::load_all("castor")
|
To install castor from the installation file on your system use:
|
install.packages("castor_X.X.X.tar.gz")
|
where "X.X.X" is the appropriate version number in the file name.
|
|
 ×
 ×
 ×
 ×
 ×
|
|
|
Louca lab. Department of Biology, University of Oregon, Eugene, USA
© 2025 Stilianos Louca all rights reserved
|
|
|



