The Libuda Lab studies how DNA is accurately repaired during sperm and egg development.


Live imaging of germ cell chromosome structures in whole intact C. elegans
How is the genome faithfully passed on through generations?
Utilizing molecular genetics, live and fixed imaging, super-resolution and high-resolution microscopy, CRISPR-Cas9 genome engineering, genomics, and biochemistry, the Libuda lab studies the molecular mechanisms behind how DNA double strand breaks (DSBs) are made and repaired during sperm and egg development to ensure genomic integrity and accurate chromosome segregation through generations.
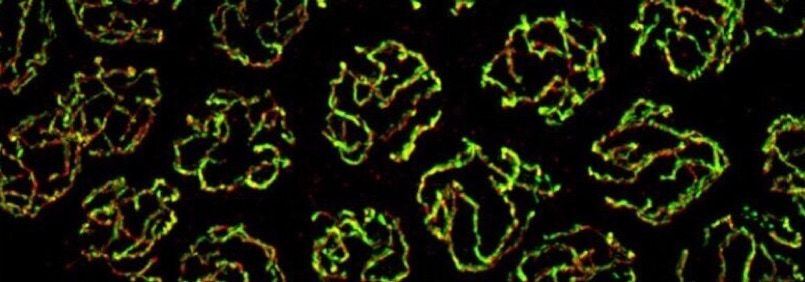
Superresolution imaging of meiotic chromosome structures
Specifically, we study DSB formation and repair during meiosis, the specialized cell division that generates sperm and eggs. For our studies, we use two model organisms that have strong conservation with human fertility processes: the nematode Caenorhabditis elegans and the zebrafish Danio rerio. Our work has strong biomedical relevance for understanding fertility issues, birth defects, and cancer.
Some of our current research interests include:
Molecular mechanisms of DNA repair pathway choice
While repair of DSBs with the appropriate chromosome template (homolog) is necessary for genomic integrity in germ cells, very little is known as to how germ cells achieve this repair template preference in the presence of other potential templates with nearly identical sequences (sister chromatids).
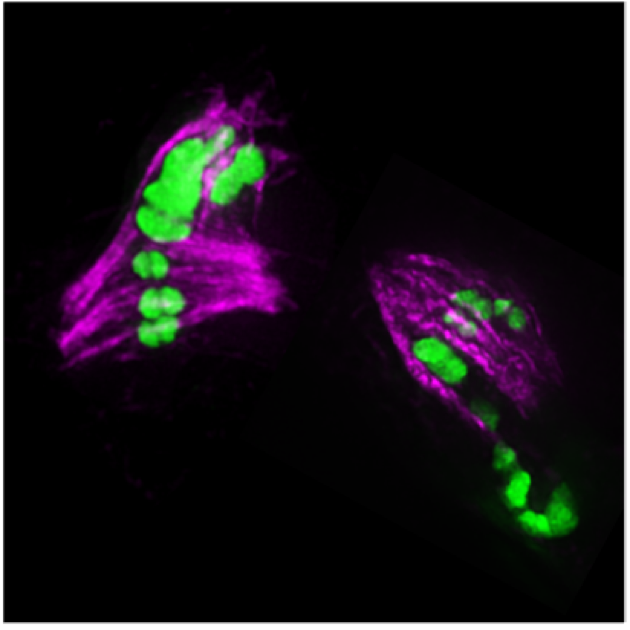
defective segregation of meiotic chromosomes (green) with excess crossovers
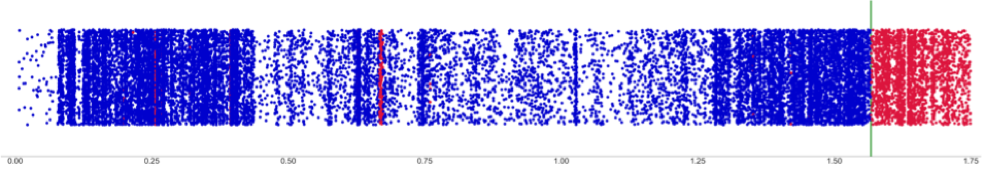
Genomic analysis of crossovers
Heat-induced male infertility
In multiple organisms from plants to humans, male fertility is acutely sensitive to changes in temperature. Our research found that acute heat shock activates mobile DNA elements, called transposons, only in developing sperm. We are uncovering the molecular mechanisms behind why only spermatogenesis is affected.
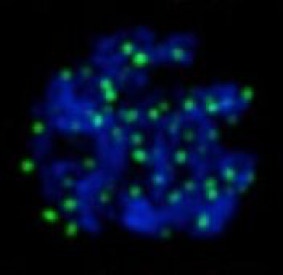
heat-induced DSBs (green) in single spermatocyte
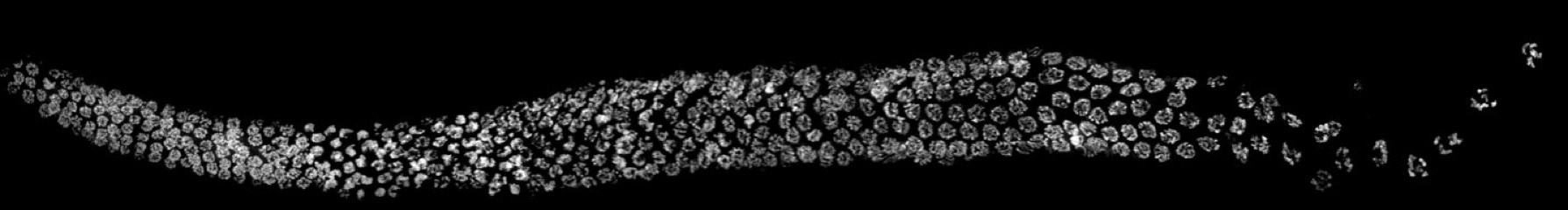
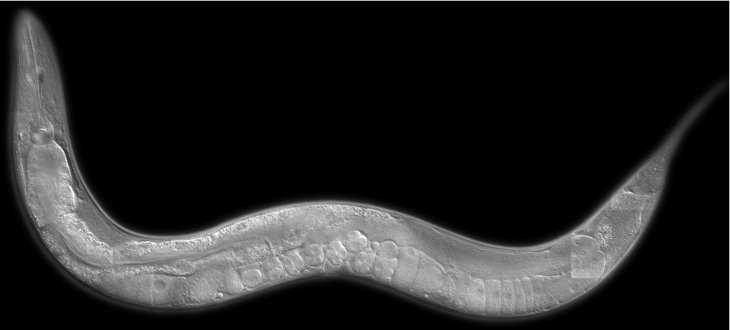
C. elegans germ line (oogenesis) stained with DNA dye DAPI
Department of Biology | Institute of Molecular Biology
For questions about the website please click here
Website last updated September 2024 by Dylan Legg